3IFX
 
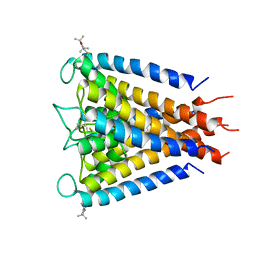 | | Crystal structure of the Spin-labeled KcsA mutant V48R1 | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, TETRABUTYLAMMONIUM ION, ... | | Authors: | Cieslak, J.A, Focia, P.J, Gross, A. | | Deposit date: | 2009-07-26 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | EPR (3.56 Å), X-RAY DIFFRACTION | | Cite: | Electron Spin-Echo Envelope Modulation (ESEEM) Reveals Water and Phosphate Interactions with the KcsA Potassium Channel
Biochemistry, 49, 2010
|
|
6AKD
 
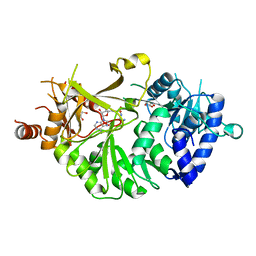 | | Crystal structure of IdnL7 | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, AMP-dependent synthetase and ligase, GLYCEROL | | Authors: | Cieslak, J, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-08-31 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of IdnL7, an adenylation enzyme involved in incednine biosynthesis.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
5JJQ
 
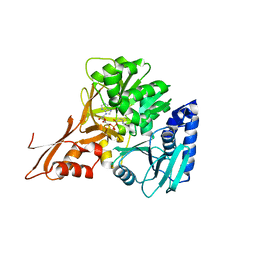 | | Crystal structure of IdnL1 | | Descriptor: | 5'-O-[(R)-{[(3S)-3-aminobutanoyl]oxy}(hydroxy)phosphoryl]adenosine, AMP-dependent synthetase and ligase, CHLORIDE ION | | Authors: | Cieslak, J, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2016-04-25 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical characterization and structural insight into aliphatic beta-amino acid adenylation enzymes IdnL1 and CmiS6
Proteins, 85, 2017
|
|
5JJP
 
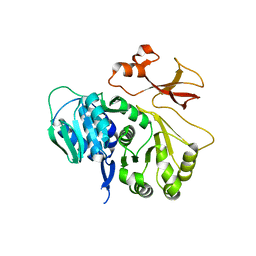 | | Crystal structure of CmiS6 | | Descriptor: | Nonribosomal peptide synthase | | Authors: | Cieslak, J, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2016-04-25 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical characterization and structural insight into aliphatic beta-amino acid adenylation enzymes IdnL1 and CmiS6
Proteins, 85, 2017
|
|
3WV5
 
 | | Complex structure of VinN with 3-methylaspartate | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WV4
 
 | | Crystal structure of VinN | | Descriptor: | Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WVN
 
 | | Complex structure of VinN with L-aspartate | | Descriptor: | ASPARTIC ACID, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-30 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
