2FMT
 
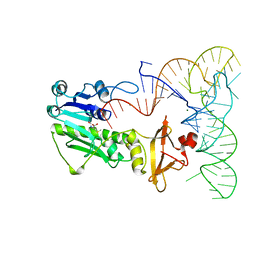 | | METHIONYL-TRNAFMET FORMYLTRANSFERASE COMPLEXED WITH FORMYL-METHIONYL-TRNAFMET | | Descriptor: | FORMYL-METHIONYL-TRNAFMET2, MAGNESIUM ION, METHIONYL-TRNA FMET FORMYLTRANSFERASE, ... | | Authors: | Schmitt, E, Mechulam, Y, Blanquet, S. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet.
EMBO J., 17, 1998
|
|
2AHO
 
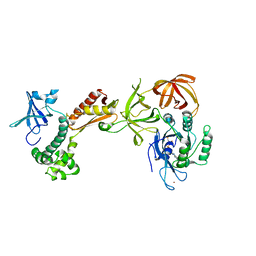 | | Structure of the archaeal initiation factor eIF2 alpha-gamma heterodimer from Sulfolobus solfataricus complexed with GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Translation initiation factor 2 alpha subunit, ... | | Authors: | Yatime, L, Mechulam, Y, Blanquet, S, Schmitt, E. | | Deposit date: | 2005-07-28 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Switch of the gamma Subunit in an Archaeal aIF2alphagamma Heterodimer
Structure, 14, 2006
|
|
9FRL
 
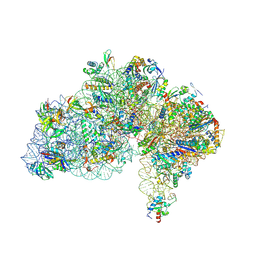 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to SD mRNA with h44 in up position | | Descriptor: | LSU ribosomal protein S30E (Rps30E), Large ribosomal subunit protein eL8, MAGNESIUM ION, ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-06-19 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9FSF
 
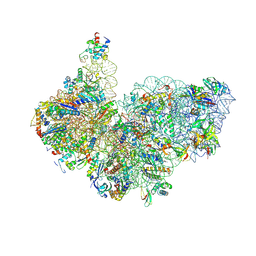 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to Ss-MAP leaderless mRNA with h44 in up position | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LSU ribosomal protein S30E (Rps30E), Large ribosomal subunit protein eL8, ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9FS8
 
 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to Ss-aEF1A-like mRNA | | Descriptor: | LSU ribosomal protein S26E (Rps26E), LSU ribosomal protein S30E (Rps30E), Large ribosomal subunit protein eL8, ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-06-20 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9FRK
 
 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to SD mRNA | | Descriptor: | LSU ribosomal protein S30E (Rps30E), Large ribosomal subunit protein eL8, MAGNESIUM ION, ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-06-19 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9FS6
 
 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to Ss-aIF2beta leaderless mRNA with h44 in up position | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, LSU ribosomal protein S26E (Rps26E), LSU ribosomal protein S30E (Rps30E), ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-06-20 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9FY0
 
 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to Ss-aIF2beta leaderless mRNA | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, LSU ribosomal protein S26E (Rps26E), LSU ribosomal protein S30E (Rps30E), ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-07-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9FRA
 
 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to Ss-MAP leaderless mRNA | | Descriptor: | LSU ribosomal protein S30E (Rps30E), Large ribosomal subunit protein eL8, MAGNESIUM ION, ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9FHL
 
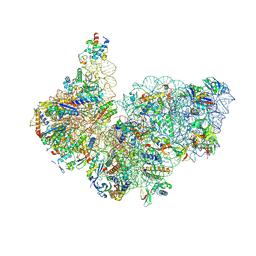 | | High-resolution cryo-EM structure of Saccharolobus solfataricus 30S ribosomal subunit bound to mRNA and initiator tRNA | | Descriptor: | Initiator tRNA Met, LSU ribosomal protein S30E (Rps30E), Large ribosomal subunit protein eL8, ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-05-27 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
3H9C
 
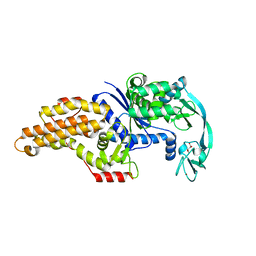 | | Structure of methionyl-tRNA synthetase: crystal form 2 | | Descriptor: | CITRIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Schmitt, E, Tanrikulu, I.C, Yoo, T.H, Panvert, M, Tirrell, D.A, Mechulam, Y. | | Deposit date: | 2009-04-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
J.Mol.Biol., 394, 2009
|
|
3H9B
 
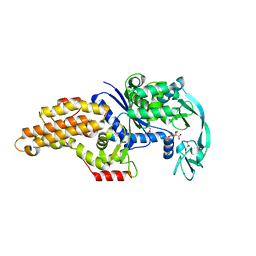 | | Structure of a mutant methionyl-tRNA synthetase with modified specificity complexed with azidonorleucine | | Descriptor: | 6-azido-L-norleucine, CITRIC ACID, Methionyl-tRNA synthetase, ... | | Authors: | Schmitt, E, Tanrikulu, I.C, Yoo, T.H, Panvert, M, Tirrell, D.A, Mechulam, Y. | | Deposit date: | 2009-04-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
J.Mol.Biol., 394, 2009
|
|
3FRH
 
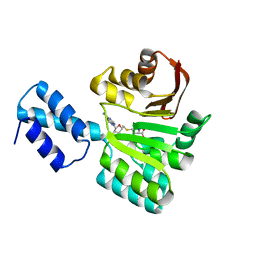 | | Structure of the 16S rRNA methylase RmtB, P21 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3H99
 
 | | Structure of a mutant methionyl-tRNA synthetase with modified specificity complexed with methionine | | Descriptor: | CITRIC ACID, METHIONINE, Methionyl-tRNA synthetase, ... | | Authors: | Schmitt, E, Tanrikulu, I.C, Yoo, T.H, Panvert, M, Tirrell, D.A, Mechulam, Y. | | Deposit date: | 2009-04-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
J.Mol.Biol., 394, 2009
|
|
3H97
 
 | | Structure of a mutant methionyl-tRNA synthetase with modified specificity | | Descriptor: | CITRIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Schmitt, E, Tanrikulu, I.C, Yoo, T.H, Panvert, M, Tirrell, D.A, Mechulam, Y. | | Deposit date: | 2009-04-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
J.Mol.Biol., 394, 2009
|
|
3FZG
 
 | | Structure of the 16S rRNA methylase ArmA | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3FRI
 
 | | Structure of the 16S rRNA methylase RmtB, I222 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
1JKE
 
 | | D-Tyr tRNATyr deacylase from Escherichia coli | | Descriptor: | D-Tyr-tRNATyr deacylase, ZINC ION | | Authors: | Ferri-Fioni, M.L, Schmitt, E, Soutourina, J, Plateau, P, Mechulam, Y, Blanquet, S. | | Deposit date: | 2001-07-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of crystalline D-Tyr-tRNA(Tyr) deacylase. A representative of a new class of tRNA-dependent hydrolases.
J.Biol.Chem., 276, 2001
|
|
1FMT
 
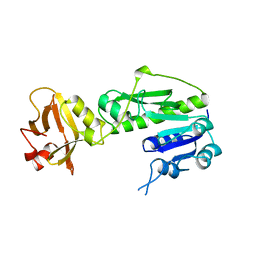 | | METHIONYL-TRNAFMET FORMYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA FMET FORMYLTRANSFERASE | | Authors: | Schmitt, E, Mechulam, Y. | | Deposit date: | 1997-10-13 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of crystalline Escherichia coli methionyl-tRNA(f)Met formyltransferase: comparison with glycinamide ribonucleotide formyltransferase.
EMBO J., 15, 1996
|
|
2QMU
 
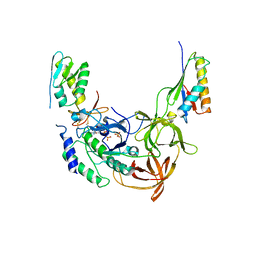 | | Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Translation initiation factor 2 alpha subunit, ... | | Authors: | Yatime, L, Mechulam, Y, Blanquet, S, Schmitt, E. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7YZN
 
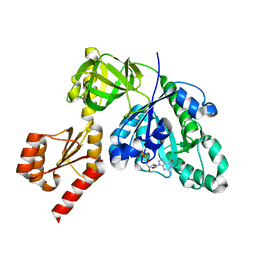 | | Structure of C-terminally truncated aIF5B from Pyrococcus abyssi complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable translation initiation factor IF-2, ... | | Authors: | Bourgeois, G, Schmitt, E, Mechulam, Y, Coureux, P.D, Kazan, R. | | Deposit date: | 2022-02-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
1P7P
 
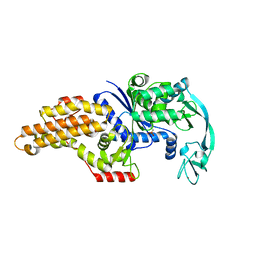 | | Methionyl-tRNA synthetase from Escherichia coli complexed with methionine phosphonate | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHONIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-05 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PFY
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH METHIONYL SULPHAMOYL ADENOSINE | | Descriptor: | 5'-O-[(L-METHIONYL)-SULPHAMOYL]ADENOSINE, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PFU
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH METHIONINE PHOSPHINATE | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PG0
 
 | | Methionyl-trna synthetase from escherichia coli complexed with methioninyl adenylate | | Descriptor: | L-METHIONYL ADENYLATE, Methionyl-tRNA synthetase | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
