3AHT
 
 | | Crystal structure of rice BGlu1 E176Q mutant in complex with laminaribiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3AHV
 
 | | Semi-active E176Q mutant of rice bglu1 covalent complex with 2-deoxy-2-fluoroglucoside | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase 7, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
2LA1
 
 | | Expression in Pichia pastoris and backbone dynamics of dendroaspin, a three finger toxin | | Descriptor: | Mambin | | Authors: | Chuang, W.J, Cheng, C.H, Chen, Y.C, Shiu, J.H. | | Deposit date: | 2011-03-01 | | Release date: | 2012-03-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dynamics and functional differences between dendroaspin and rhodostomin: Insights into protein scaffolds in integrin recognition
Protein Sci., 21, 2012
|
|
3SZG
 
 | | Crystal structure of C176A glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi and NaAD+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T, Gerratana, B. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
3SYT
 
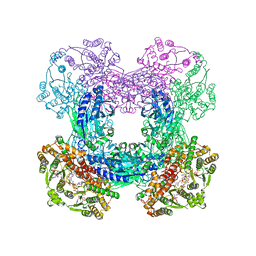 | | Crystal structure of glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi, NAD+, and glutamate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Chuenchor, W, Gerratana, B. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6511 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
2M75
 
 | |
2M7H
 
 | |
2M7F
 
 | |
2MB3
 
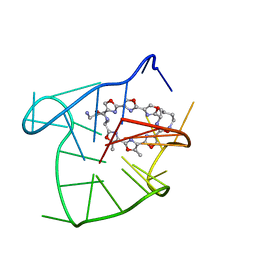 | | Solution structure of an intramolecular (3+1) human telomeric G-quadruplex bound to a telomestatin derivative | | Descriptor: | (12S,27S)-12,27-bis(4-aminobutyl)-4,30-dimethyl-3,7,14,18,22,29-hexaoxa-11,26,31,32,33,34,35,36-octaazaheptacyclo[26.2. 1.1~2,5~.1~6,9~.1~13,16~.1~17,20~.1~21,24~]hexatriaconta-1(30),2(36),4,6(35),8,13(34),15,17(33),19,21(32),23,28(31)-dode caene-10,25-dione, DNA_(5'-D(*TP*TP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*A)-3') | | Authors: | Chung, W.J, Heddi, B, Tera, M, Iida, K, Nagasawa, K, Phan, A.T. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular (3 + 1) human telomeric g-quadruplex bound to a telomestatin derivative.
J.Am.Chem.Soc., 135, 2013
|
|
2MGN
 
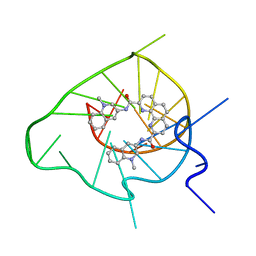 | | Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3 | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3', N2,N9-bis(1-methylquinolin-3-yl)-1,10-phenanthroline-2,9-dicarboxamide | | Authors: | Chung, W.J, Heddi, B, Hamon, F, Teulade-Fichou, M.P, Phan, A.T. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a G-quadruplex Bound to the Bisquinolinium Compound Phen-DC3.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3F5K
 
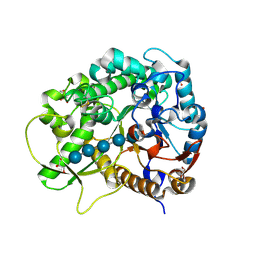 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F5J
 
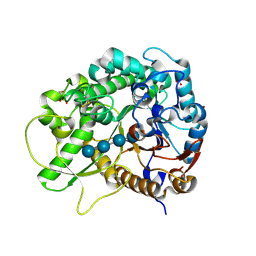 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
1H6T
 
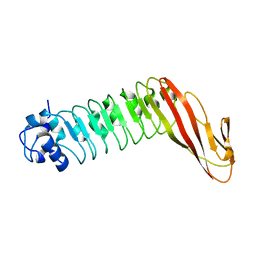 | | Internalin B: crystal structure of fused N-terminal domains. | | Descriptor: | INTERNALIN B | | Authors: | Schubert, W.-D, Gobel, G, Diepholz, M, Darji, A, Kloer, D, Hain, T, Chakraborty, T, Wehland, J, Domann, E, Heinz, D.W. | | Deposit date: | 2001-06-22 | | Release date: | 2001-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain.
J.Mol.Biol., 312, 2001
|
|
2PJI
 
 | |
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F4V
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
1A6B
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | Authors: | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | Deposit date: | 1998-02-23 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
3PCY
 
 | | THE CRYSTAL STRUCTURE OF MERCURY-SUBSTITUTED POPLAR PLASTOCYANIN AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | MERCURY (II) ION, PLASTOCYANIN | | Authors: | Church, W.B, Guss, J.M, Potter, J.J, Freeman, H.C. | | Deposit date: | 1985-12-10 | | Release date: | 1986-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of mercury-substituted poplar plastocyanin at 1.9-A resolution.
J.Biol.Chem., 261, 1986
|
|
1H6U
 
 | | Internalin H: crystal structure of fused N-terminal domains. | | Descriptor: | INTERNALIN H | | Authors: | Schubert, W.-D, Gobel, G, Diepholz, M, Darji, A, Kloer, D, Hain, T, Chakraborty, T, Wehland, J, Domann, E, Heinz, D.W. | | Deposit date: | 2001-06-25 | | Release date: | 2001-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain.
J.Mol.Biol., 312, 2001
|
|
1Q7J
 
 | |
1Q7I
 
 | |
1IBI
 
 | | QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, 15 MINIMIZED MODEL STRUCTURES | | Descriptor: | CYSTEINE-RICH PROTEIN 2, ZINC ION | | Authors: | Schuler, W, Kloiber, K, Matt, T, Bister, K, Konrat, R. | | Deposit date: | 2001-03-28 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Application of cross-correlated NMR spin relaxation to the zinc-finger protein CRP2(LIM2): evidence for collective motions in LIM domains.
Biochemistry, 40, 2001
|
|
3SEQ
 
 | |
3SEZ
 
 | |
3SDB
 
 | |
