5GL6
 
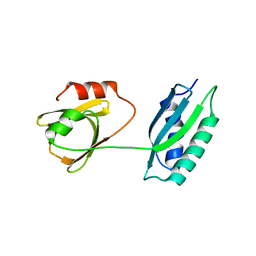 | | Msmeg rimP | | Descriptor: | Ribosome maturation factor RimP | | Authors: | Chu, T, Lau, T.C.K. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ribosomal maturation factor P fromMycobacterium smegmatisfacilitates the ribosomal biogenesis by binding to the small ribosomal protein S12.
J. Biol. Chem., 294, 2019
|
|
6QAM
 
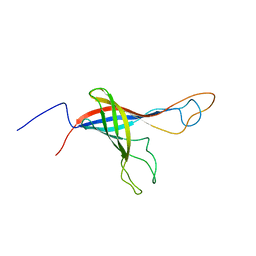 | |
3HJ6
 
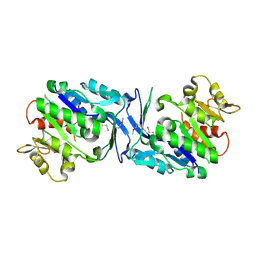 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
6QWR
 
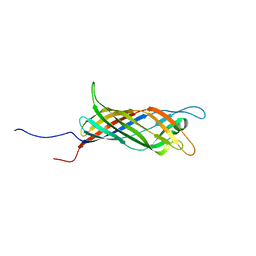 | |
8TLS
 
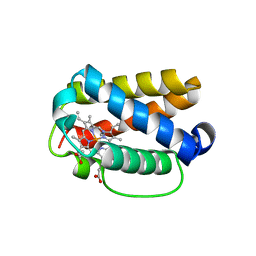 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y108A variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y108A variant
To Be Published
|
|
8UGZ
 
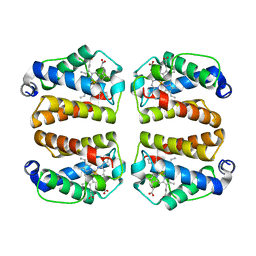 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant
To Be Published
|
|
8UZU
 
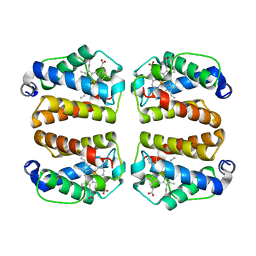 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin L80A C51S C71S variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin L80A C51S C71S variant
To Be Published
|
|
3IIS
 
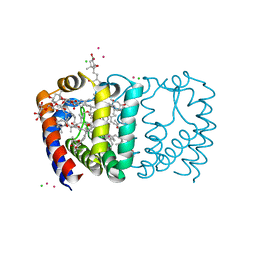 | | Structure of the reconstituted Peridinin-Chlorophyll a-Protein (RFPCP) | | Descriptor: | (2S)-3-[(6-O-alpha-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-2-[(3Z,6Z,9Z,12Z,15Z)-octadeca-3,6,9,12,15-pentaenoyloxy]propyl (5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Schulte, T, Hofmann, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of a single peridinin sensing Chl-a excitation in reconstituted PCP by crystallography and spectroscopy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6TA7
 
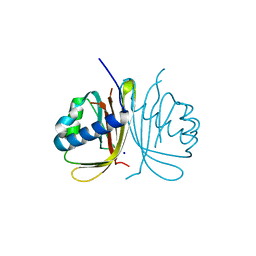 | | CRYSTAL STRUCTURE OF HUMAN G3BP1-NTF2 IN COMPLEX WITH HUMAN CAPRIN1-DERIVED SOLOMON MOTIF | | Descriptor: | CHLORIDE ION, Caprin-1, Ras GTPase-activating protein-binding protein 1, ... | | Authors: | Schulte, T, Achour, A, Panas, M.D, McInerney, G.M. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Caprin-1 binding to the critical stress granule protein G3BP1 is regulated by pH
Biorxiv, 2021
|
|
3T2N
 
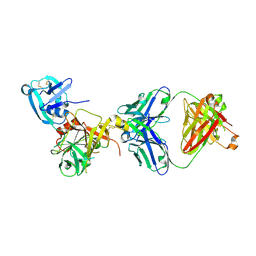 | | Human hepsin protease in complex with the Fab fragment of an inhibitory antibody | | Descriptor: | Antibody, Fab fragment, Heavy Chain, ... | | Authors: | Koschubs, T, Dengl, S, Duerr, H, Kaluza, K, Georges, G, Hartl, C, Jennewein, S, Lanzendoerfer, M, Auer, J, Stern, A, Huang, K.-S, Kostrewa, D, Ries, S, Hansen, S, Kohnert, U, Cramer, P, Mundigl, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric antibody inhibition of human hepsin protease.
Biochem.J., 442, 2012
|
|
3IIU
 
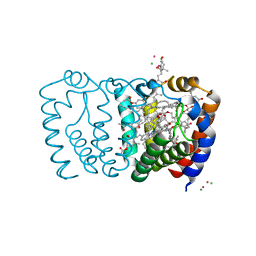 | | Structure of the reconstituted Peridinin-Chlorophyll a-Protein (RFPCP) mutant N89L | | Descriptor: | (2S)-3-[(6-O-alpha-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-2-[(3Z,6Z,9Z,12Z,15Z)-octadeca-3,6,9,12,15-pentaenoyloxy]propyl (5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Schulte, T, Hofmann, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of a single peridinin sensing Chl-a excitation in reconstituted PCP by crystallography and spectroscopy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5JUI
 
 | |
5FW5
 
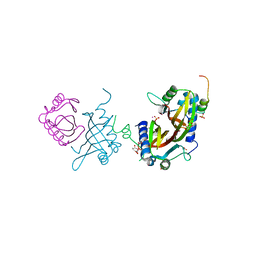 | | Crystal structure of human G3BP1 in complex with Semliki Forest Virus nsP3-25 comprising two FGDF motives | | Descriptor: | ACETATE ION, GLYCEROL, NON-STRUCTURAL PROTEIN 3, ... | | Authors: | Schulte, T, Liu, L, Panas, M.D, Thaa, B, Goette, B, Achour, A, McInerney, G.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication.
Open Biol, 6, 2016
|
|
8TPE
 
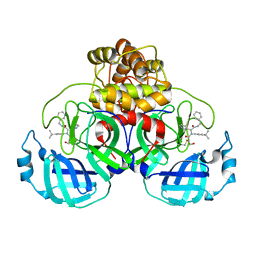 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPB
 
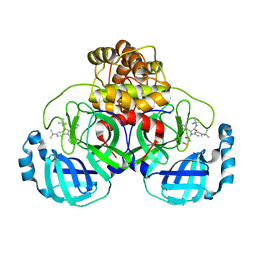 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-2-chloroacetamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPG
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPH
 
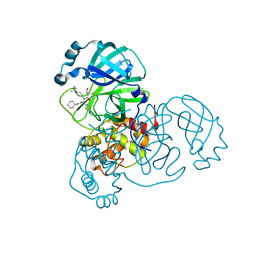 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPI
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-2-hydroxy-2-methylpropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPD
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | 3C-like proteinase nsp5, N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[3-(2-chloroacetamido)phenyl]furan-2-carboxamide | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPF
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPC
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[4-(2-chloroacetamido)phenyl]furan-2-carboxamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
5WF9
 
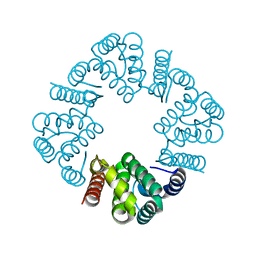 | | Tepsin tENTH domain 1-153 | | Descriptor: | AP-4 complex accessory subunit Tepsin | | Authors: | Archuleta, T.L, Jackson, L.P. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and evolution of ENTH and VHS/ENTH-like domains in tepsin.
Traffic, 18, 2017
|
|
5WFB
 
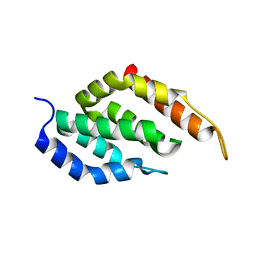 | | Tepsin tENTH domain 1-136. | | Descriptor: | AP-4 complex accessory subunit Tepsin | | Authors: | Archuleta, T.L, Jackson, L.P. | | Deposit date: | 2017-07-11 | | Release date: | 2017-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Structure and evolution of ENTH and VHS/ENTH-like domains in tepsin.
Traffic, 18, 2017
|
|
7ZH7
 
 | |
2X1Z
 
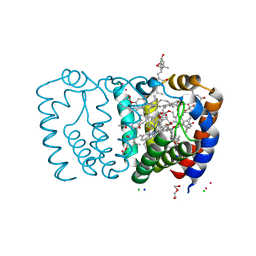 | | Structure of Peridinin-Chlorophyll-Protein reconstituted with Chl-d | | Descriptor: | CADMIUM ION, CHLORIDE ION, CHLOROPHYLL D, ... | | Authors: | Schulte, T, Hiller, R.G, Hofmann, E. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Peridinin-Chlorophyll-Protein Reconstituted with Different Chlorophylls.
FEBS Lett., 584, 2010
|
|
