3PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI IN COMPLEX WITH SUBSTRATE ANALOGUE OXAMATE | | Descriptor: | OXAMIC ACID, PROTEIN (FORMATE ACETYLTRANSFERASE 1) | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
2PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI | | Descriptor: | CHLORIDE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-26 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
2EZI
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, 30 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
1CM5
 
 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | Descriptor: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1RDR
 
 | | POLIOVIRUS 3D POLYMERASE | | Descriptor: | CALCIUM ION, POLIOVIRUS 3D POLYMERASE | | Authors: | Hansen, J, Long, A, Schultz, S. | | Deposit date: | 1998-04-28 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase of poliovirus.
Structure, 5, 1997
|
|
1OTC
 
 | | THE O. NOVA TELOMERE END BINDING PROTEIN COMPLEXED WITH SINGLE STRAND DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), PROTEIN (TELOMERE-BINDING PROTEIN ALPHA SUBUNIT), PROTEIN (TELOMERE-BINDING PROTEIN BETA SUBUNIT) | | Authors: | Horvath, M.P, Schweiker, V.L, Bevilacqua, J.M, Ruggles, J.A, Schultz, S.C. | | Deposit date: | 1998-11-25 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA.
Cell(Cambridge,Mass.), 95, 1998
|
|
1PH8
 
 | |
1PH9
 
 | |
1PH6
 
 | |
1PHJ
 
 | |
1PH1
 
 | |
1PH2
 
 | |
1PH3
 
 | |
1PH5
 
 | |
1PH4
 
 | |
1PH7
 
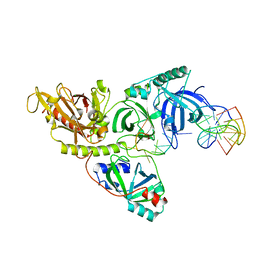 | |
2EZK
 
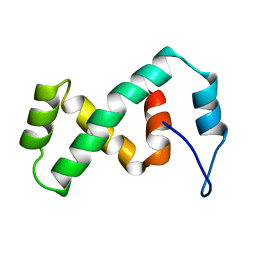 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, REGULARIZED MEAN STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZL
 
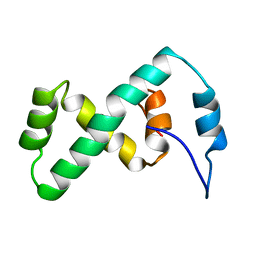 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, 29 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
1DI2
 
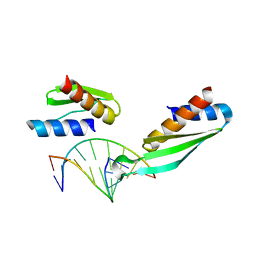 | |
1PA6
 
 | |
2OB0
 
 | | Human MAK3 homolog in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Human MAK3 homolog | | Authors: | Walker, J.R, Schuetz, S, Antoshenko, T, Wu, H, Bernstein, G, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-18 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human MAK3 homolog
To be Published
|
|
1HDQ
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with D-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HDU
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HEE
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with L-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, L-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
