8U4C
 
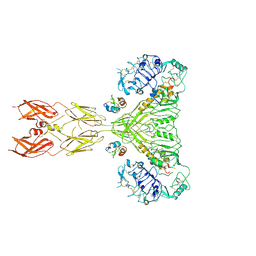 | | Cryo-EM structure of long form insulin receptor (IR-B) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
7TYJ
 
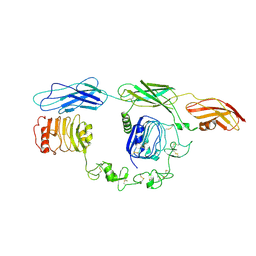 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was focused on one of two halves with C1 symmetry applied | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYK
 
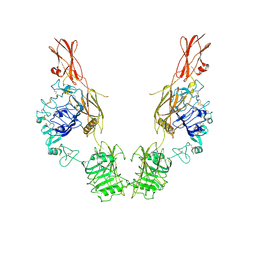 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYM
 
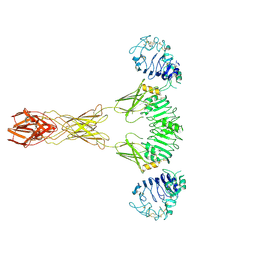 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in active-state captured at pH 9. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
