1YGM
 
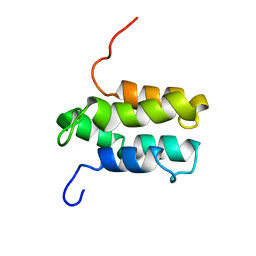 | | NMR structure of Mistic | | Descriptor: | hypothetical protein BSU31320 | | Authors: | Roosild, T.P, Greenwald, J, Vega, M, Castronovo, S, Riek, R, Choe, S. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mistic, a membrane-integrating protein for membrane protein expression.
Science, 307, 2005
|
|
1EOF
 
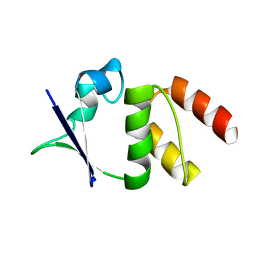 | | CRYSTAL STRUCTURE OF THE N136A MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
1A68
 
 | |
1B8K
 
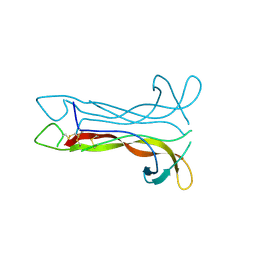 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B8M
 
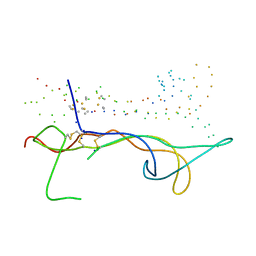 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | Descriptor: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1LSS
 
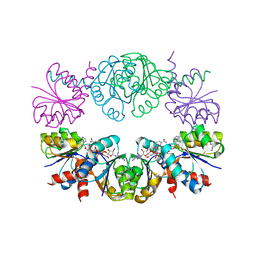 | | KTN Mja218 CRYSTAL STRUCTURE IN COMPLEX WITH NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Trk system potassium uptake protein trkA homolog | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
1B98
 
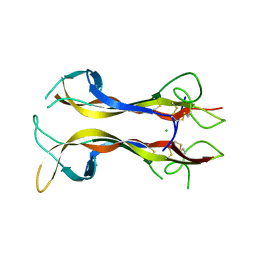 | | NEUROTROPHIN 4 (HOMODIMER) | | Descriptor: | CHLORIDE ION, PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-22 | | Release date: | 1999-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B9F
 
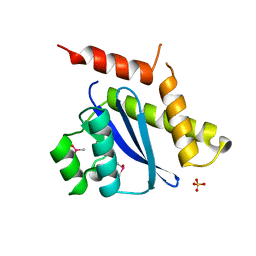 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B92
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-19 | | Release date: | 1999-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B9D
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1BTE
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF THE TYPE II ACTIVIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (ACTIVIN RECEPTOR TYPE II) | | Authors: | Greenwald, J, Fischer, W, Vale, W, Choe, S. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase.
Nat.Struct.Biol., 6, 1999
|
|
1LSU
 
 | | KTN Bsu222 Crystal Structure in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Conserved hypothetical protein yuaA | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
1HYV
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH TETRAPHENYL ARSONIUM | | Descriptor: | CHLORIDE ION, INTEGRASE, SULFATE ION, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HYZ
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH A DERIVATIVE OF TETRAPHENYL ARSONIUM. | | Descriptor: | (3,4-DIHYDROXY-PHENYL)-TRIPHENYL-ARSONIUM, CHLORIDE ION, INTEGRASE, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1COS
 
 | | CRYSTAL STRUCTURE OF A SYNTHETIC TRIPLE-STRANDED ALPHA-HELICAL BUNDLE | | Descriptor: | COILED SERINE | | Authors: | Lovejoy, B, Choe, S, Cascio, D, Mcrorie, D.K, Degrado, W, Eisenberg, D. | | Deposit date: | 1993-01-22 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a synthetic triple-stranded alpha-helical bundle.
Science, 259, 1993
|
|
1U4E
 
 | | Crystal Structure of Cytoplasmic Domains of GIRK1 channel | | Descriptor: | G protein-activated inward rectifier potassium channel 1 | | Authors: | Pegan, S, Arrabit, C, Zhou, W, Kwiatkowski, W, Slesinger, P.A, Choe, S. | | Deposit date: | 2004-07-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification
Nat.Neurosci., 8, 2005
|
|
1U4F
 
 | | Crystal Structure of Cytoplasmic Domains of IRK1 (Kir2.1) channel | | Descriptor: | Inward rectifier potassium channel 2 | | Authors: | Pegan, S, Arrabit, C, Zhou, W, Kwiatkowski, W, Slesinger, P.A, Choe, S. | | Deposit date: | 2004-07-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification
Nat.Neurosci., 8, 2005
|
|
1OMT
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
2GOO
 
 | | Ternary Complex of BMP-2 bound to BMPR-Ia-ECD and ActRII-ECD | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Activin receptor type 2A, Bone morphogenetic protein 2, ... | | Authors: | Allendorph, G.P, Choe, S. | | Deposit date: | 2006-04-13 | | Release date: | 2006-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the ternary signaling complex of a TGF-beta superfamily member.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2KSE
 
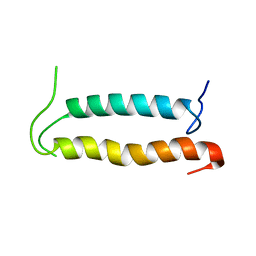 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor QseC, Center for Structures of Membrane Proteins (CSMP) target 4311C | | Descriptor: | Sensor protein qseC | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KSD
 
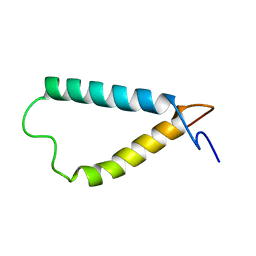 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor ArcB, Center for Structures of Membrane Proteins (CSMP) target 4310C | | Descriptor: | Aerobic respiration control sensor protein arcB | | Authors: | Maslennikov, I, Klammt, C, Hwang, E, Kefala, G, Kwiatkowski, W, Jeon, Y, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KSF
 
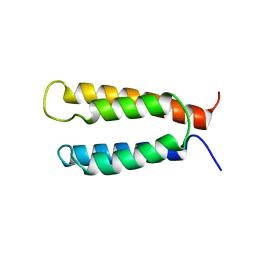 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor KdpD, Center for Structures of Membrane Proteins (CSMP) target 4312C | | Descriptor: | Sensor protein kdpD | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Okamura, M, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-03 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LON
 
 | |
2LOR
 
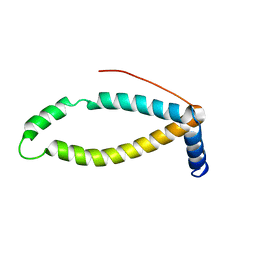 | | Backbone structure of human membrane protein TMEM141 | | Descriptor: | Transmembrane protein 141 | | Authors: | Bayrhuber, M, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOO
 
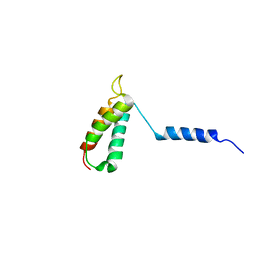 | | Backbone structure of human membrane protein TMEM14A from NOE data | | Descriptor: | Transmembrane protein 14A | | Authors: | Eichmann, C, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
