1A68
 
 | |
1B98
 
 | | NEUROTROPHIN 4 (HOMODIMER) | | Descriptor: | CHLORIDE ION, PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-22 | | Release date: | 1999-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
3Q41
 
 | | Crystal structure of the GluN1 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit zeta-1 | | Authors: | Farina, A.N, Blain, K.Y, Maruo, T, Kwiatkowski, W, Choe, S, Nakagawa, T. | | Deposit date: | 2010-12-22 | | Release date: | 2011-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Separation of Domain Contacts Is Required for Heterotetrameric Assembly of Functional NMDA Receptors.
J.Neurosci., 31, 2011
|
|
1B8K
 
 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1T1D
 
 | |
2KSE
 
 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor QseC, Center for Structures of Membrane Proteins (CSMP) target 4311C | | Descriptor: | Sensor protein qseC | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2MOI
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MOL
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2GOO
 
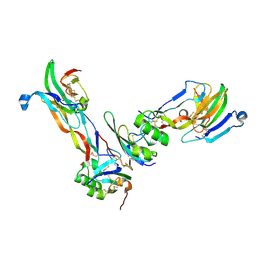 | | Ternary Complex of BMP-2 bound to BMPR-Ia-ECD and ActRII-ECD | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Activin receptor type 2A, Bone morphogenetic protein 2, ... | | Authors: | Allendorph, G.P, Choe, S. | | Deposit date: | 2006-04-13 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the ternary signaling complex of a TGF-beta superfamily member.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5LAM
 
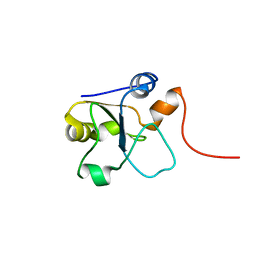 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
2KSD
 
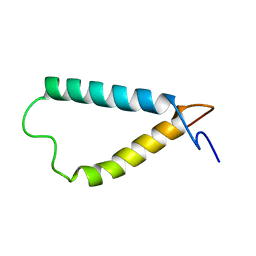 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor ArcB, Center for Structures of Membrane Proteins (CSMP) target 4310C | | Descriptor: | Aerobic respiration control sensor protein arcB | | Authors: | Maslennikov, I, Klammt, C, Hwang, E, Kefala, G, Kwiatkowski, W, Jeon, Y, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1YAE
 
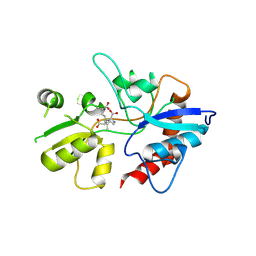 | | Structure of the Kainate Receptor Subunit GluR6 Agonist Binding Domain Complexed with Domoic Acid | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor, ... | | Authors: | Nanao, M.H, Green, T, Stern-Bach, Y, Heinemann, S.F, Choe, S. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
5LAO
 
 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
1BTE
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF THE TYPE II ACTIVIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (ACTIVIN RECEPTOR TYPE II) | | Authors: | Greenwald, J, Fischer, W, Vale, W, Choe, S. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase.
Nat.Struct.Biol., 6, 1999
|
|
1B8M
 
 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | Descriptor: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
2KSF
 
 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor KdpD, Center for Structures of Membrane Proteins (CSMP) target 4312C | | Descriptor: | Sensor protein kdpD | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Okamura, M, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-03 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LOM
 
 | | Backbone structure of human membrane protein HIGD1A | | Descriptor: | HIG1 domain family member 1A | | Authors: | Blain, K, Klammt, C, Maslennikov, I, Kwiatkowski, W, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOS
 
 | | Backbone structure of human membrane protein TMEM14C | | Descriptor: | Transmembrane protein 14C | | Authors: | Klammt, C, Vajpai, N, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOQ
 
 | | Backbone structure of human membrane protein FAM14B (Interferon alpha-inducible protein 27-like protein 1) | | Descriptor: | Interferon alpha-inducible protein 27-like protein 1 | | Authors: | Klammt, C, Chui, E.J, Maslennikov, I, Kwiatkowski, W, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
1EOD
 
 | | CRYSTAL STRUCTURE OF THE N136D MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
1EOE
 
 | | CRYSTAL STRUCTURE OF THE V135R MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
1EOF
 
 | | CRYSTAL STRUCTURE OF THE N136A MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
2MPN
 
 | | 3D NMR structure of the transmembrane domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-05-29 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
1ZKZ
 
 | | Crystal Structure of BMP9 | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Brown, M.A, Zhao, Q, Baker, K.A, Naik, C, Chen, C, Pukac, L, Singh, M, Tsareva, T, Parice, Y, Mahoney, A, Roschke, V, Sanyal, I, Choe, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of BMP-9 and functional interactions with pro-region and receptors
J.Biol.Chem., 280, 2005
|
|
