2FEB
 
 | | NMR Solution Structure, Dynamics and Binding Properties of the Kringle IV Type 8 module of apolipoprotein(a) | | Descriptor: | Apolipoprotein(a) | | Authors: | Chitayat, S, Kanelis, V, Koschinsky, M.L, Smith, S.P. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance (NMR) solution structure, dynamics, and binding properties of the kringle IV type 8 module of apolipoprotein(a).
Biochemistry, 46, 2007
|
|
2JH2
 
 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
2O4E
 
 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
2JNK
 
 | |
2KZR
 
 | | Solution NMR Structure of Ubiquitin thioesterase OTU1 (EC 3.1.2.-) from Mus musculus, Northeast Structural Genomics Consortium Target MmT2A | | Descriptor: | Ubiquitin thioesterase OTU1 | | Authors: | Chitayat, S, Gutmanas, A, Lemak, A, Yee, A, Bezsonova, I, Wu, B, Doherty, R.S, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target MmT2A
To be Published
|
|
2LTJ
 
 | | Conformational analysis of StrH, the surface-attached exo- beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Pluvinage, B, Chitayat, S, Ficko-Blean, E, Abbott, D, Kunjachen, J, Grondin, J, Spencer, H, Smith, S, Boraston, A. | | Deposit date: | 2012-05-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational analysis of StrH, the surface-attached exo-beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae.
J.Mol.Biol., 425, 2013
|
|
2KWF
 
 | | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1 | | Descriptor: | CREB-binding protein, Transcription factor 4 | | Authors: | Denis, C.M, Chitayat, S, Plevin, M.J, Liu, S, Spencer, H.L, Ikura, M, LeBrun, D.P, Smith, S.P. | | Deposit date: | 2010-04-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1
To be Published
|
|
2M1U
 
 | | Solution structure of the small dictyostelium discoideium myosin light chain mlcb provides insights into iq-motif recognition of class i myosin myo1b | | Descriptor: | myosin light chain MlcB | | Authors: | Liburd, J, Chitayat, S, Crawley, S.W, Denis, C.M, Cote, G.P, Smith, S.P. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the small Dictyostelium discoideum myosin light chain MlcB provides insights into MyoB IQ motif recognition.
J.Biol.Chem., 289, 2014
|
|
2LS6
 
 | | Solution NMR Structure of a Non-canonical galactose-binding CBM32 from Clostridium perfringens | | Descriptor: | Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Chitayat, S, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2012-04-20 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An unusual mode of galactose recognition by a family 32 carbohydrate-binding module.
J.Mol.Biol., 426, 2014
|
|
2M8U
 
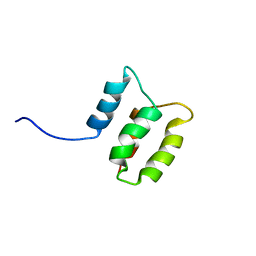 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | Descriptor: | Myosin Light Chain, MlcC | | Authors: | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | Deposit date: | 2013-05-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2MH0
 
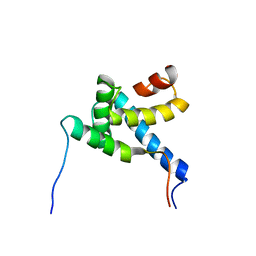 | |
2LFE
 
 | | Solution NMR structure of N-terminal domain of human E3 ubiquitin-protein ligase HECW2, Northeast structural genomics consortium (NESG) target ht6306A | | Descriptor: | E3 ubiquitin-protein ligase HECW2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Chitayat, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-29 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of E3 ligase HECW2
To be Published
|
|
