8IN9
 
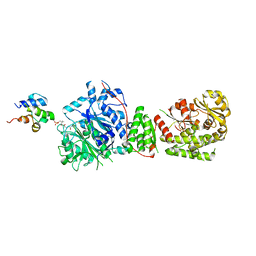 | | The structure of the GfsA KSQ-AT didomain in complex with the GfsA ACP domain | | 分子名称: | N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, Polyketide synthase | | 著者 | Chisuga, T, Murakami, S, Miyanaga, A, Kudo, F, Eguchi, T. | | 登録日 | 2023-03-09 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure-Based Analysis of Transient Interactions between Ketosynthase-like Decarboxylase and Acyl Carrier Protein in a Loading Module of Modular Polyketide Synthase.
Acs Chem.Biol., 18, 2023
|
|
7VEE
 
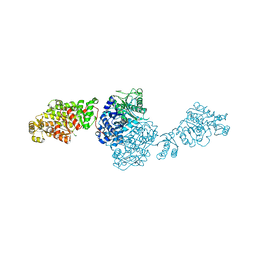 | | The ligand-free structure of GfsA KSQ-AT didomain | | 分子名称: | GLYCEROL, Polyketide synthase | | 著者 | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | 登録日 | 2021-09-08 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
7VEF
 
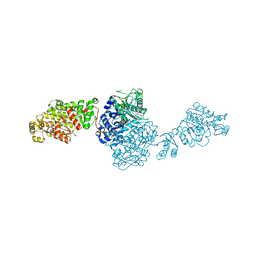 | | The structure of GfsA KSQ-AT didomain in complex with a malonate substrate analog | | 分子名称: | GLYCEROL, N-(2-acetamidoethyl)-2-nitro-ethanamide, Polyketide synthase | | 著者 | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | 登録日 | 2021-09-08 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
5WSX
 
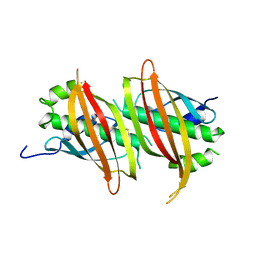 | | The crystal structure of SAV606 | | 分子名称: | Uncharacterized protein | | 著者 | Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | 登録日 | 2016-12-08 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analysis of the dual-function thioesterase SAV606 unravels the mechanism of Michael addition of glycine to an alpha , beta-unsaturated thioester.
J. Biol. Chem., 292, 2017
|
|
5WSY
 
 | | The complex structure of SAV606 with N-carboxymethyl-3-aminobutyrate | | 分子名称: | (3~{R})-3-(2-hydroxy-2-oxoethylamino)butanoic acid, Uncharacterized protein | | 著者 | Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | 登録日 | 2016-12-08 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of the dual-function thioesterase SAV606 unravels the mechanism of Michael addition of glycine to an alpha , beta-unsaturated thioester.
J. Biol. Chem., 292, 2017
|
|
8H6S
 
 | | Structure of acyltransferase VinK in complex with the loading acyl carrier protein of vicenistatin PKS | | 分子名称: | MAGNESIUM ION, Malonyl-CoA-[acyl-carrier-protein] transacylase, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | 著者 | Kawada, K, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | 登録日 | 2022-10-18 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis of Transient Interactions of Acyltransferase VinK with the Loading Acyl Carrier Protein of the Vicenistatin Modular Polyketide Synthase.
Biochemistry, 62, 2023
|
|
8K4R
 
 | | Structure of VinM-VinL complex | | 分子名称: | Acyl-carrier-protein, Non-ribosomal peptide synthetase, SODIUM ION, ... | | 著者 | Miyanaga, A, Nagata, K, Nakajima, J, Chisuga, T, Kudo, F, Eguchi, T. | | 登録日 | 2023-07-20 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of Amide-Forming Adenylation Enzyme VinM in Vicenistatin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8YYQ
 
 | | Structure of the HitB F328L mutant | | 分子名称: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(3-cyanophenyl)propanoyl]sulfamate | | 著者 | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | 登録日 | 2024-04-04 | | 公開日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 2024
|
|
8YYR
 
 | | Structure of the HitB T293G mutant | | 分子名称: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(2-bromophenyl)propanoyl]sulfamate | | 著者 | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | 登録日 | 2024-04-04 | | 公開日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 2024
|
|
6K97
 
 | | Crystal structure of fusion DH domain | | 分子名称: | Fusion DH, SULFATE ION | | 著者 | Kawasaki, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | 登録日 | 2019-06-14 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional and Structural Analyses of the Split-Dehydratase Domain in the Biosynthesis of Macrolactam Polyketide Cremimycin.
Biochemistry, 58, 2019
|
|
6J38
 
 | | Crystal structure of CmiS2 | | 分子名称: | FAD-dependent glycine oxydase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Kawasaki, D, Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | 登録日 | 2019-01-04 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analysis of the Glycine Oxidase Homologue CmiS2 Reveals a Unique Substrate Recognition Mechanism for Formation of a beta-Amino Acid Starter Unit in Cremimycin Biosynthesis.
Biochemistry, 58, 2019
|
|
6J39
 
 | | Crystal structure of CmiS2 with inhibitor | | 分子名称: | (3R)-3-[(carboxymethyl)sulfanyl]nonanoic acid, FAD-dependent glycine oxydase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Kawasaki, D, Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | 登録日 | 2019-01-04 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Analysis of the Glycine Oxidase Homologue CmiS2 Reveals a Unique Substrate Recognition Mechanism for Formation of a beta-Amino Acid Starter Unit in Cremimycin Biosynthesis.
Biochemistry, 58, 2019
|
|
