7K5U
 
 | | Bst DNA polymerase I time-resolved structure, 48 hr post dATP and dCTP addition | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*C)-3'), DNA (5'-D(P*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*C)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
7K5P
 
 | | Bst DNA polymerase I time-resolved structure, 4 min post dATP addition | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
7K5S
 
 | | Bst DNA polymerase I time-resolved structure, 4 hr post dATP and dCTP addition | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*C)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
7K5O
 
 | | Bst DNA polymerase I time-resolved structure, 1 min post dATP addition | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
7K5Q
 
 | | Bst DNA polymerase I time-resolved structure, 8 min post dATP addition | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(P*G*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
7K5R
 
 | | Bst DNA polymerase I time-resolved structure, 120 min post dATP addition | | Descriptor: | DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
7K5T
 
 | | Bst DNA polymerase I time-resolved structure, 25.5 hr post dATP and dCTP addition | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*C)-3'), ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
1PGY
 
 | | Solution structure of the UBA domain in Saccharomyces cerevisiae protein, Swa2p | | Descriptor: | Swa2p | | Authors: | Chim, N, Gall, W.E, Xiao, J, Harris, M.P, Graham, T.R, Krezel, A.M. | | Deposit date: | 2003-05-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-binding domain in Swa2p from Saccharomyces cerevisiae.
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3IOS
 
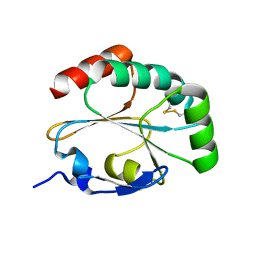 | | Structure of MTB dsbF in its mixed oxidized and reduced forms | | Descriptor: | Disulfide bond forming protein (DsbF) | | Authors: | Chim, N, Goulding, C.W. | | Deposit date: | 2009-08-14 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An extracellular disulfide bond forming protein (DsbF) from Mycobacterium tuberculosis: structural, biochemical, and gene expression analysis.
J.Mol.Biol., 396, 2010
|
|
5VU5
 
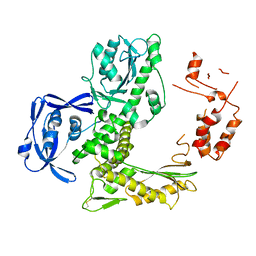 | | TNA polymerase, apo | | Descriptor: | DNA polymerase | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
6DSV
 
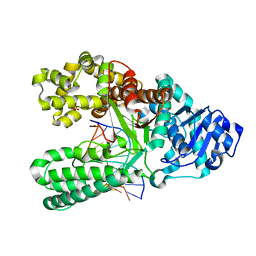 | | Bst DNA polymerase I post-chemistry (n+2) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
3HX9
 
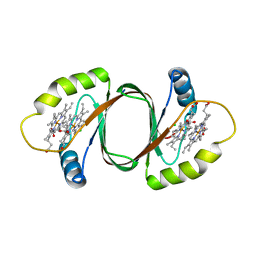 | | Structure of heme-degrader, MhuD (Rv3592), from Mycobacterium tuberculosis with two hemes bound in its active site | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Protein Rv3592 | | Authors: | Chim, N, Nguyen, T.Q, Iniguez, A, Goulding, C.W. | | Deposit date: | 2009-06-19 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unusual Diheme Conformation of the Heme-Degrading Protein from Mycobacterium tuberculosis
J.Mol.Biol., 395, 2009
|
|
6DSW
 
 | | Bst DNA polymerase I pre-chemistry (n) structure | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*G)-3'), DNA (5'-D(P*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
5VU7
 
 | | TNA polymerase, open ternary complex | | Descriptor: | 5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXYTETRAHYDROFURAN-3-YL DIHYDROGEN PHOSPHATE, DNA polymerase, DNA template, ... | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
6DSY
 
 | | Bst DNA polymerase I post-chemistry (n+1) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
5VU9
 
 | | TNA polymerase, translocated product | | Descriptor: | DNA polymerase, DNA template, DNA/TNA hybrid primer | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
5VU8
 
 | | TNA polymerase, closed ternary complex | | Descriptor: | DNA polymerase, DNA template, DNA/TNA hybrid primer, ... | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
6DSU
 
 | | Bst DNA polymerase I pre-insertion complex structure | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
5VU6
 
 | |
6DSX
 
 | | Bst DNA polymerase I post-chemistry (n+1 with dATP soak) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
5SLA
 
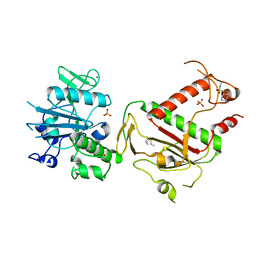 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1003207278 | | Descriptor: | 1-cyclohexyl-N-methylmethanesulfonamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLP
 
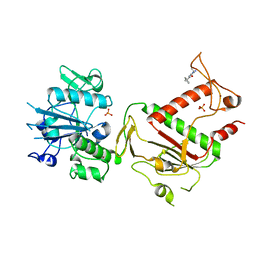 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z373768898 | | Descriptor: | N-(1-ethyl-1H-pyrazol-4-yl)cyclopentanecarboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL6
 
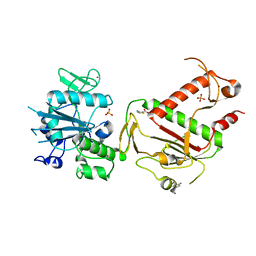 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z256709556 | | Descriptor: | 3-methylthiophene-2-carboxylic acid, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLK
 
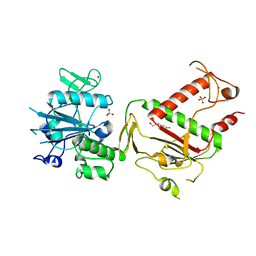 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1354370680 | | Descriptor: | 2-[(5-chloro-3-fluoropyridin-2-yl)(methyl)amino]ethan-1-ol, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLS
 
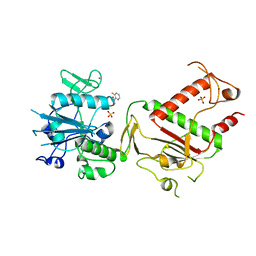 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1373445602 | | Descriptor: | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
