4HYE
 
 | |
3UYU
 
 | | Structural basis for the antifreeze activity of an ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, GLYCEROL | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
3UYV
 
 | | Crystal structure of a glycosylated ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
3E0O
 
 | | Crystal structure of MsrB | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Park, A.K, Shin, Y.J, Kim, Y.K, Chi, Y.M, Hwang, K.Y. | | Deposit date: | 2008-07-31 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Kinetic Analysis of an MsrA-MsrB Fusion Protein from Streptococcus pneumoniae
Mol.Microbiol., 72, 2009
|
|
2ZHZ
 
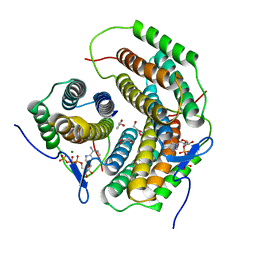 | | Crystal structure of a pduO-type ATP:cobalamin adenosyltransferase from Burkholderia thailandensis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP:cob(I)alamin adenosyltransferase, putative, ... | | Authors: | Moon, J.H, Park, A.K, Jang, E.H, Kim, H.S, Chi, Y.M. | | Deposit date: | 2008-02-12 | | Release date: | 2008-07-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a PduO-type ATP:cobalamin adenosyltransferase from Burkholderia thailandensis.
Proteins, 72, 2008
|
|
2ZHY
 
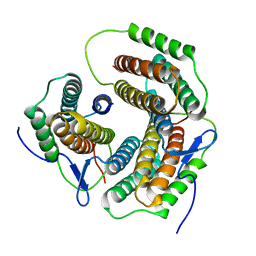 | | Crystal structure of a pduO-type ATP:cobalamin adenosyltransferase from Burkholderia thailandensis | | Descriptor: | ATP:cob(I)alamin adenosyltransferase, putative | | Authors: | Moon, J.H, Park, A.K, Jang, E.H, Kim, H.S, Chi, Y.M. | | Deposit date: | 2008-02-11 | | Release date: | 2008-07-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a PduO-type ATP:cobalamin adenosyltransferase from Burkholderia thailandensis.
Proteins, 72, 2008
|
|
