1WUH
 
 | | Three-Dimensional Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis for the activation process of [NiFe] hydrogenase from D.vulgaris Miyazaki F
To be Published
|
|
5XED
 
 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
1QGI
 
 | | CHITOSANASE FROM BACILLUS CIRCULANS | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CHITOSANASE), SULFATE ION | | Authors: | Saito, J, Kita, A, Higuchi, Y, Nagata, Y, Ando, A, Miki, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism.
J.Biol.Chem., 274, 1999
|
|
1WUK
 
 | | High resolution Structure Of The Oxidized State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
6ABU
 
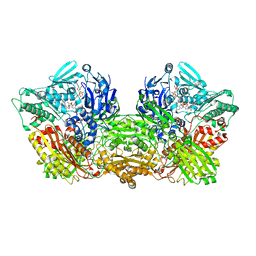 | | Rat Xanthine oxidoreductase, NAD bound form | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Rat Xanthine oxidoreductase, NAD bound form
To Be Published
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
5XVC
 
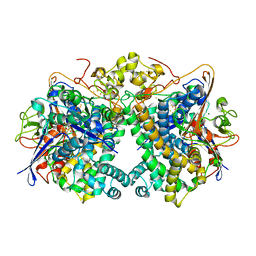 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XEC
 
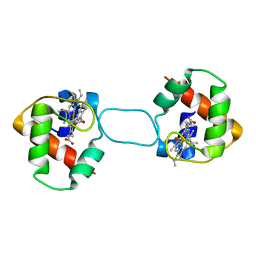 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5XLF
 
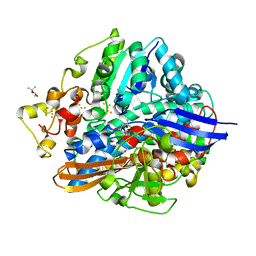 | | Crystal structure of aerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
1CPQ
 
 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS CAPSULATA | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1995-08-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | High-resolution crystal structures of two polymorphs of cytochrome c' from the purple phototrophic bacterium rhodobacter capsulatus.
J.Mol.Biol., 259, 1996
|
|
2Z1T
 
 | |
7VU6
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 3 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Yamamoto, S, Yamane, J, Tachibana, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
1WUL
 
 | | High Resolution Structure Of The Reduced State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
7VTH
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 2-[4-[[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]amino]-2,6-bis(oxidanylidene)-3-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazin-1-yl]-N-methyl-ethanamide, 3C-like proteinase | | Authors: | Yamamoto, S, Tachibana, Y. | | Deposit date: | 2021-10-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
5XLG
 
 | | Crystal structure of anaerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
1WYG
 
 | | Crystal Structure of a Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S) | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Hori, H, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2005-02-14 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Conversion of Xanthine Dehydrogenase to Xanthine Oxidase: IDENTIFICATION OF THE TWO CYSTEINE DISULFIDE BONDS AND CRYSTAL STRUCTURE OF A NON-CONVERTIBLE RAT LIVER XANTHINE DEHYDROGENASE MUTANT
J.Biol.Chem., 280, 2005
|
|
5XLH
 
 | | Crystal structure of aerobically purified and aerobically crystallized for 12weeks D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLE
 
 | | Crystal structure of anaerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XVB
 
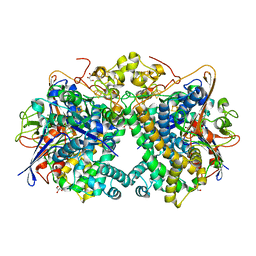 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
2ZM8
 
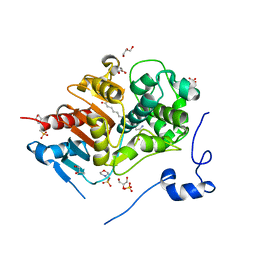 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/D370Y Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
1V66
 
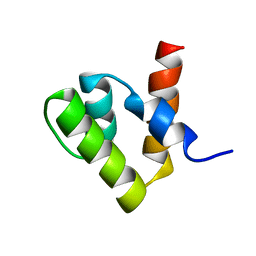 | | Solution structure of human p53 binding domain of PIAS-1 | | Descriptor: | Protein inhibitor of activated STAT protein 1 | | Authors: | Okubo, S, Hara, F, Tsuchida, Y, Shimotakahara, S, Suzuki, S, Hatanaka, H, Yokoyama, S, Tanaka, H, Yasuda, H, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers
J.Biol.Chem., 279, 2004
|
|
2Z1U
 
 | |
1WUI
 
 | | Ultra-High resolution Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1V9F
 
 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
