8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | 分子名称: | GLYCEROL, alpha/beta hydrolase | | 著者 | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | 登録日 | 2022-10-24 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
5ZE2
 
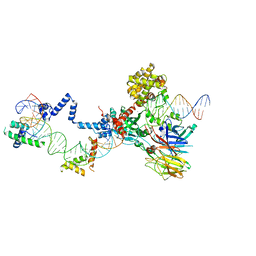 | | Hairpin Complex, RAG1/2-hairpin 12RSS/23RSS complex in 5mM Mn2+ for 2 min at 4'C | | 分子名称: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (31-MER), ... | | 著者 | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | 登録日 | 2018-02-25 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZDZ
 
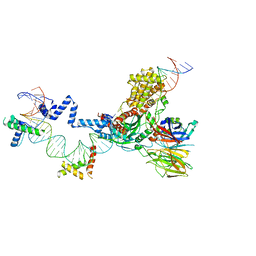 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in Ca2+ | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (30-MER), ... | | 著者 | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | 登録日 | 2018-02-25 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZE0
 
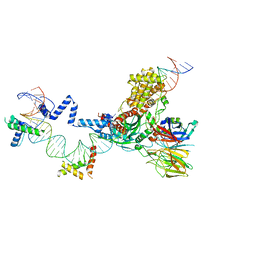 | | Hairpin Forming Complex, RAG1/2-Nicked(with Dideoxy) 12RSS/23RSS complex in Mg2+ | | 分子名称: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (39-MER), ... | | 著者 | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | 登録日 | 2018-02-25 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZE1
 
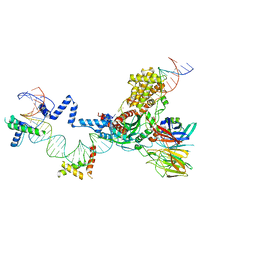 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in 2mM Mn2+ for 10 min at 4'C | | 分子名称: | 1,2-ETHANEDIOL, DNA, HMGB1 A-B box, ... | | 著者 | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | 登録日 | 2018-02-25 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
7ET0
 
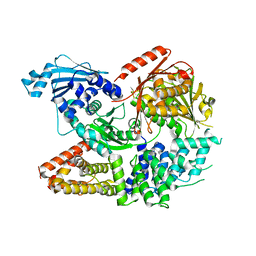 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB from wPip | | 分子名称: | Bacteria factor A, Bacteria factor B | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-05-12 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESX
 
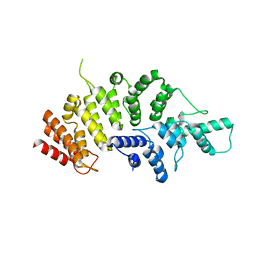 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wPip | | 分子名称: | Bacteria factor 1 | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-05-12 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESZ
 
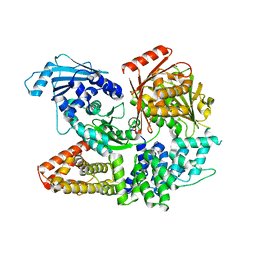 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB with Mn2+ from wPip | | 分子名称: | BACTERIA FACTOR A, BACTERIA FACTOR B, MANGANESE (II) ION | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-05-12 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.476 Å) | | 主引用文献 | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESY
 
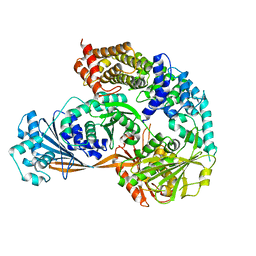 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip | | 分子名称: | Bacteria factor 1, CALCIUM ION, ULP_PROTEASE domain-containing protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-05-12 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6JPJ
 
 | | Crystal structure of FGF401 in complex of FGFR4 | | 分子名称: | Fibroblast growth factor receptor 4, N-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-7-methanoyl-6-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide, SULFATE ION | | 著者 | Zhou, Z, Chen, X, Chen, Y. | | 登録日 | 2019-03-27 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.638 Å) | | 主引用文献 | Characterization of FGF401 as a reversible covalent inhibitor of fibroblast growth factor receptor 4.
Chem.Commun.(Camb.), 55, 2019
|
|
4NKJ
 
 | |
4R37
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | 著者 | Fisher, A.J, Chen, X, Ngo, A. | | 登録日 | 2014-08-14 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R83
 
 | | Crystal structure of Sialyltransferase from Photobacterium damsela | | 分子名称: | CALCIUM ION, Sialyltransferase 0160 | | 著者 | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | 登録日 | 2014-08-29 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R36
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | 著者 | Ngo, A, Fong, K, Cox, D, Fisher, A, Chen, X. | | 登録日 | 2014-08-14 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R9V
 
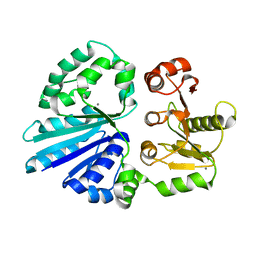 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | 分子名称: | CALCIUM ION, Sialyltransferase 0160 | | 著者 | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | 登録日 | 2014-09-08 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R84
 
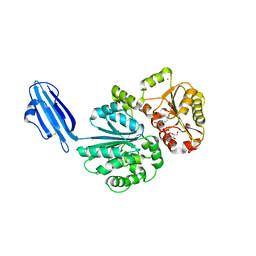 | | Crystal structure of Sialyltransferase from Photobacterium damsela with CMP-3F(a)Neu5Ac bound | | 分子名称: | CALCIUM ION, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, Sialyltransferase 0160 | | 著者 | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | 登録日 | 2014-08-29 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
7YCX
 
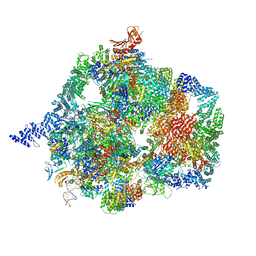 | | The structure of INTAC-PEC complex | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | 登録日 | 2022-07-02 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | 分子名称: | CHLORIDE ION, SODIUM ION, Vps75 | | 著者 | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | 登録日 | 2021-09-10 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
6L03
 
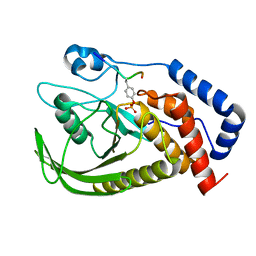 | | structure of PTP-MEG2 and MUNC18-1-pY145 peptide complex | | 分子名称: | Tyrosine-protein phosphatase non-receptor type 9, stxbp1-pY145 peptide | | 著者 | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | 登録日 | 2019-09-25 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.084 Å) | | 主引用文献 | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
6KZQ
 
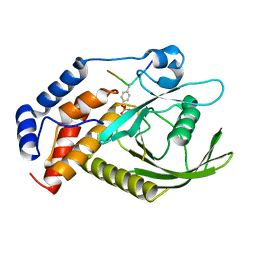 | | structure of PTP-MEG2 and NSF-pY83 peptide complex | | 分子名称: | NSF-pY83 peptide, Tyrosine-protein phosphatase non-receptor type 9 | | 著者 | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | 登録日 | 2019-09-25 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
5ZHY
 
 | | Structural characterization of the HCoV-229E fusion core | | 分子名称: | Spike glycoprotein | | 著者 | Zhang, W, Zheng, Q, Yan, M, Chen, X, Yang, H, Zhou, W, Rao, Z. | | 登録日 | 2018-03-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.441 Å) | | 主引用文献 | Structural characterization of the HCoV-229E fusion core.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
6L6H
 
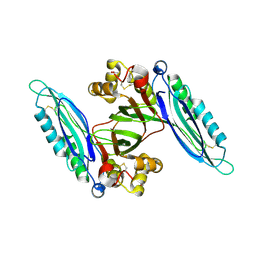 | | Crystal structure of Lpg0189 | | 分子名称: | ACETATE ION, GLYCEROL, Uncharacterized protein Lpg0189 | | 著者 | Ge, H, Chen, X. | | 登録日 | 2019-10-29 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.403 Å) | | 主引用文献 | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6L6G
 
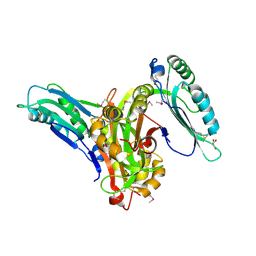 | | Crystal structure of SeMet_Lpg0189 | | 分子名称: | GLYCEROL, Uncharacterized protein Lpg0189 | | 著者 | Ge, H, Chen, X. | | 登録日 | 2019-10-29 | | 公開日 | 2019-11-27 | | 最終更新日 | 2020-01-15 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.393 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.919 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
