4ETP
 
 | | C-terminal motor and motor homology domain of Kar3Vik1 fused to a synthetic heterodimeric coiled coil | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Kinesin-like protein KAR3, ... | | Authors: | Rank, K.C, Chen, C.J, Cope, J, Porche, K, Hoenger, A, Gilbert, S.P, Rayment, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kar3Vik1, a member of the kinesin-14 superfamily, shows a novel kinesin microtubule binding pattern.
J.Cell Biol., 197, 2012
|
|
4HEQ
 
 | | The crystal structure of flavodoxin from Desulfovibrio gigas | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Dimeric Flavodoxin from Desulfovibrio gigas Suggests a Potential Binding Region for the Electron-Transferring Partner
Int J Mol Sci, 14, 2013
|
|
6KBL
 
 | | Structure-function study of AKR4C14, an aldo-keto reductase from Thai Jasmine rice (Oryza sativa L. ssp. Indica cv. KDML105) | | Descriptor: | ACETATE ION, Aldo-keto reductase, CACODYLATE ION, ... | | Authors: | Songsiriritthigul, C, Narawongsanont, R, Guan, H.H, Chen, C.J. | | Deposit date: | 2019-06-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function study of AKR4C14, an aldo-keto reductase from Thai jasmine rice (Oryza sativa L. ssp. indica cv. KDML105).
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | Authors: | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
6L0I
 
 | | Crystal structure of dihydroorotase in complex with malate at pH6.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0F
 
 | | Crystal structure of dihydroorotase in complex with 5-Aminouracil from Saccharomyces cerevisiae | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0K
 
 | | Crystal structure of dihydroorotase in complex with malate at pH9 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
6L0J
 
 | | Crystal structure of Dihydroorotase in complex with malate at pH7.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism
Molecules, 2021
|
|
6L0H
 
 | | Crystal structure of dihydroorotase in complex with malate at pH7 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
7WQU
 
 | | FeoC from Klebsiella pneumoniae | | Descriptor: | Ferrous iron transport protein B, Probable [Fe-S]-dependent transcriptional repressor | | Authors: | Hsueh, K.L, Yu, L.K, Hsieh, Y.C, Hsiao, Y.Y, Chen, C.J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.202 Å) | | Cite: | FeoC from Klebsiella pneumoniae uses its iron sulfur cluster to regulate the GTPase activity of the ferrous iron channel.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
5XGT
 
 | |
5XSK
 
 | | Crystal structure of PWWP-DNA complex for human hepatoma-derived growth factor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*TP*GP*GP*TP*CP*TP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*AP*AP*GP*AP*CP*CP*A)-3'), ... | | Authors: | Chen, L.Y, Huang, Y.C, Hsieh, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2017-06-14 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of PWWP-DNA complex at 2.84 Angstroms resolution
To Be Published
|
|
5XSL
 
 | |
7CFW
 
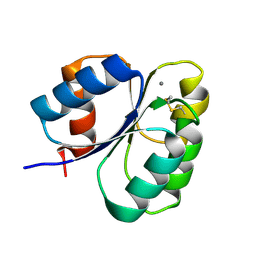 | | Crystal structure of the receiver domain of sensor histidine kinase PA1611 (PA1611REC) from Pseudomonas aeruginosa PAO1 with calcium ion coordinated in the active site cleft | | Descriptor: | CALCIUM ION, Histidine kinase | | Authors: | Chen, S.K, Guan, H.H, Wu, P.H, Lin, L.T, Wu, M.C, Chang, H.Y, Chen, N.C, Lin, C.C, Chuankhayan, P, Huang, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Iucrj, 7, 2020
|
|
3PTM
 
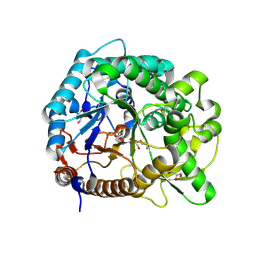 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 with 2-fluoroglucopyranoside | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase Os4BGlu12, GLYCEROL, ... | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
3PTK
 
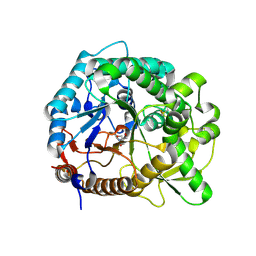 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase Os4BGlu12, ZINC ION | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
3OR1
 
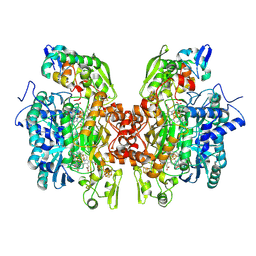 | | Crystal structure of dissimilatory sulfite reductase I (DsrI) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure insights into the enzyme catalysis from comparison of three forms of dissimilatory sulfite reductase from Desulfovibrio gigas
To be Published
|
|
3PTQ
 
 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 with dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, Beta-glucosidase Os4BGlu12, GLYCEROL, ... | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
3OR2
 
 | | Crystal structure of dissimilatory sulfite reductase II (DsrII) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissimilatory Sulfite Reductase, Sulfate Reduction
To be Published
|
|
7CDL
 
 | | holo-methanol dehydrogenase (MDH) with Cys131-Cys132 reduced from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-20 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CE9
 
 | | PQQ-soaked Apo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CFX
 
 | | NAD-soaked Holo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CED
 
 | | Apo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, large subunit | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CE5
 
 | | Methanol-PQQ bound methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, METHANOL, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7EXF
 
 | | Crystal structure of wild-type from Arabidopsis thaliana complexed with Galactose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
