1M32
 
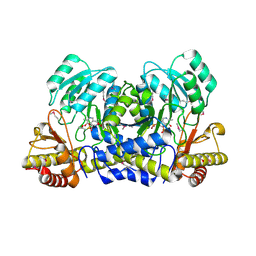 | | Crystal Structure of 2-aminoethylphosphonate Transaminase | | Descriptor: | 2-aminoethylphosphonate-pyruvate aminotransferase, PHOSPHATE ION, PHOSPHONOACETALDEHYDE, ... | | Authors: | Chen, C.C.H, Zhang, H, Kim, A.D, Howard, A, Sheldrick, G.M, Mariano-Dunnaway, D, Herzberg, O. | | Deposit date: | 2002-06-26 | | Release date: | 2002-11-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Degradation Pathway of the Phosphonate Ciliatine: Crystal Structure of 2-Aminoethylphosphonate Transaminase
Biochemistry, 41, 2002
|
|
2HJP
 
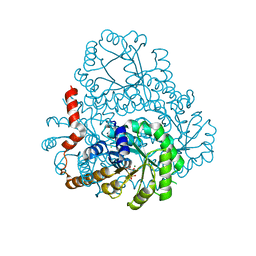 | |
2HRW
 
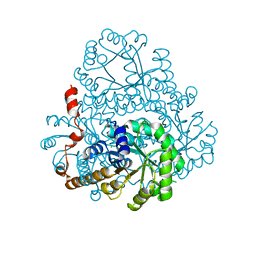 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
1JOE
 
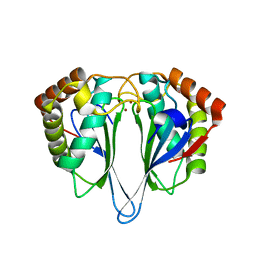 | | Crystal Structure of Autoinducer-2 Production Protein (LuxS) from Heamophilus influenzae | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Chen, C.C.H, Parsons, J.F, Lim, K, Lehmann, C, Tempczyk, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-27 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF AUTOINDUCER-2 PRODUCTION PROTEIN (LUXS) FROM HEAMOPHILUS INFLUENZAE--A CASE OF TWINNED CRYSTAL
To be Published
|
|
1BLH
 
 | |
1BLC
 
 | |
1KGF
 
 | |
1KGE
 
 | |
1KGG
 
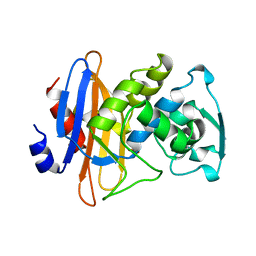 | | STRUCTURE OF BETA-LACTAMASE GLU166GLN:ASN170ASP MUTANT | | Descriptor: | PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relocation of the catalytic carboxylate group in class A beta-lactamase: the structure and function of the mutant enzyme Glu166-->Gln:Asn170-->Asp.
Protein Eng., 12, 1999
|
|
1DJC
 
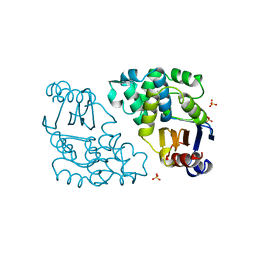 | |
1DJA
 
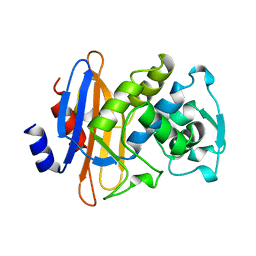 | |
1DJB
 
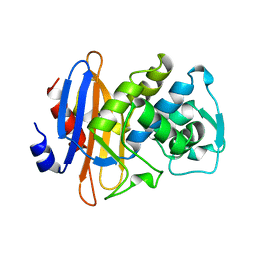 | |
1GHM
 
 | | Structures of the acyl-enzyme complex of the staphylococcus aureus beta-lactamase mutant GLU166ASP:ASN170GLN with degraded cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, BETA-LACTAMASE, CARBONATE ION, ... | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2000-12-19 | | Release date: | 2001-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of the acyl-enzyme complexes of the Staphylococcus aureus beta-lactamase mutant Glu166Asp:Asn170Gln with benzylpenicillin and cephaloridine.
Biochemistry, 40, 2001
|
|
1GHP
 
 | |
1GHI
 
 | | STRUCTURE OF BETA-LACTAMASE GLU166ASP:ASN170GLN MUTANT | | Descriptor: | BETA-LACTAMASE, CARBONATE ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2000-12-18 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the acyl-enzyme complexes of the Staphylococcus aureus beta-lactamase mutant Glu166Asp:Asn170Gln with benzylpenicillin and cephaloridine.
Biochemistry, 40, 2001
|
|
1DIK
 
 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
