1S2J
 
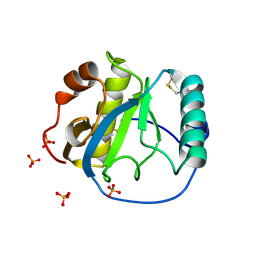 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
1C4R
 
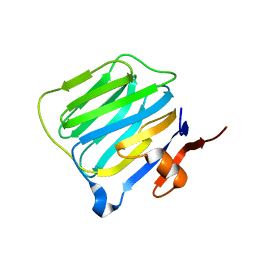 | | THE STRUCTURE OF THE LIGAND-BINDING DOMAIN OF NEUREXIN 1BETA: REGULATION OF LNS DOMAIN FUNCTION BY ALTERNATIVE SPLICING | | Descriptor: | NEUREXIN-I BETA | | Authors: | Rudenko, G, Nguyen, T, Chelliah, Y, Sudhof, T.C, Deisenhofer, J. | | Deposit date: | 1999-09-28 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing.
Cell(Cambridge,Mass.), 99, 1999
|
|
6PTZ
 
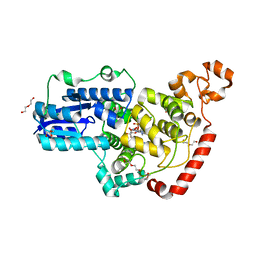 | | Crystal structure of pigeon Cryptochrome 4 mutant Y319D in complex with flavin adenine dinucleotide | | Descriptor: | Cryptochrome-1, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zoltowski, B.D, Chelliah, Y, Wickramaratne, A.C, Jarocha, L, Karki, N, Mouritsen, H, Hore, P.J, Hibbs, R.E, Green, C.B, Takahashi, J.S. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4F3L
 
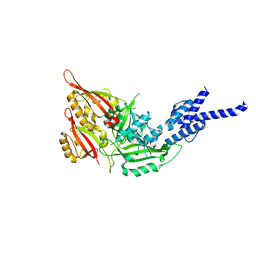 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
6PU0
 
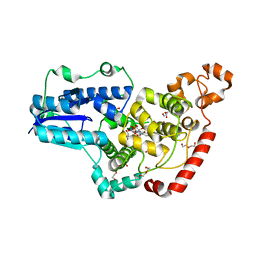 | | Pigeon Cryptochrome4 bound to flavin adenine dinucleotide | | Descriptor: | 1,2-ETHANEDIOL, Cryptochrome-1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zoltowski, B.D, Chelliah, Y, Wickramaratne, A.C, Jarocha, L, Karki, N, Mouritsen, H, Hore, P.J, Hibbs, R.E, Green, C.B, Takahashi, J.S. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8979 Å) | | Cite: | Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3O6N
 
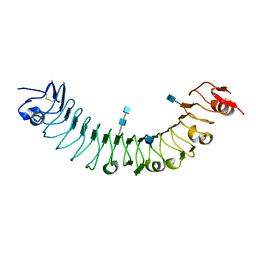 | | Crystal Structure of APL1 leucine-rich repeat domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, APL1 | | Authors: | Baxter, R.H.G, Steinert, S, Chelliah, Y, Volohonsky, G, Levashina, E.A, Deisenhofer, J. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1PX2
 
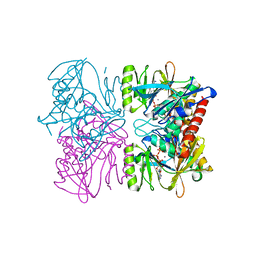 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP (Form 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Synapsin I | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-07-02 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
1PK8
 
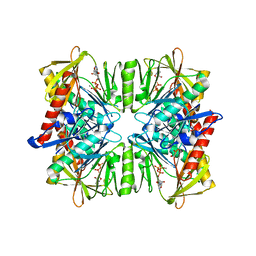 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-06-05 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
3O53
 
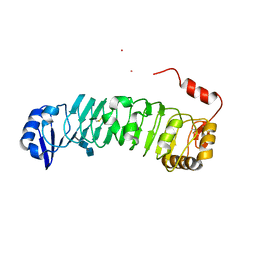 | | Crystal Structure of LRIM1 leucine-rich repeat domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Baxter, R.H.G, Steinert, S, Chelliah, Y, Volohonsky, G, Levashina, E.A, Deisenhofer, J. | | Deposit date: | 2010-07-27 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IO4
 
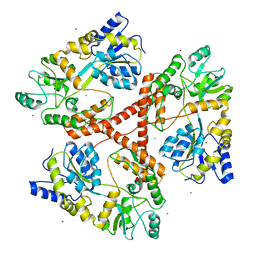 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IO6
 
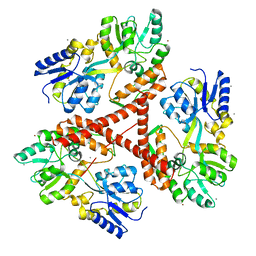 | | Huntingtin amino-terminal region with 17 Gln residues - crystal C92-a | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein, HUNTINGTIN FUSION PROTEIN, ... | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
1ZZV
 
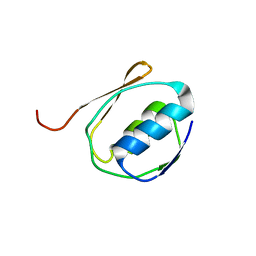 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter FecA from Escherichia coli. | | Descriptor: | Iron(III) dicitrate transport protein fecA | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-14 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
2A02
 
 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | Descriptor: | Ferric-pseudobactin 358 receptor | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-15 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
1Z6I
 
 | | Crystal structure of the ectodomain of Drosophila transmembrane receptor PGRP-LCa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidoglycan-recognition protein-LC, SULFATE ION | | Authors: | Chang, C.-I, Ihara, K, Chelliah, Y, Mengin-Lecreulx, D, Wakatsuki, S, Deisenhofer, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A4H
 
 | | Solution structure of Sep15 from Drosophila melanogaster | | Descriptor: | Selenoprotein Sep15 | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
2A2P
 
 | | Solution structure of SelM from Mus musculus | | Descriptor: | Selenoprotein M | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
