3GIN
 
 | |
2AK7
 
 | | structure of a dimeric P-Ser-Crh | | Descriptor: | HPr-like protein crh, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Poncet, S, Lecampion, C, Lariviere, L, Meyer, P, Galinier, A, Deutscher, J, Nessler, S, Morera, S. | | Deposit date: | 2005-08-03 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a domain-swapped dimer of Ser46-phosphorylated Crh from Bacillus subtilis.
Proteins, 63, 2006
|
|
2FEP
 
 | | Structure of truncated CcpA in complex with P-Ser-HPr and Sulfate ions | | Descriptor: | Catabolite control protein A, Phosphocarrier protein HPr, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Poncet, S, Lecampion, C, Meyer, P, Deutscher, J, Galinier, A, Nessler, S. | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of B. subtilis CcpA effector binding site.
Proteins, 64, 2006
|
|
2GAA
 
 | |
2GA8
 
 | |
6R72
 
 | | Crystal structure of BmrA-E504A in an outward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug exporter ATP-binding cassette | | Authors: | Chaptal, V, Zampieri, V, Kilburg, A, Magnard, S, Falson, P. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
2QMH
 
 | | structure of V267F mutant HprK/P | | Descriptor: | HPr kinase/phosphorylase | | Authors: | Chaptal, V, Vincent, F, Gueguen-Chaignon, V, Poncet, S, Deutscher, J, Nessler, S, Morera, S. | | Deposit date: | 2007-07-16 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Bacterial HPr Kinase/Phosphorylase V267F Mutant Gives Insights into the Allosteric Regulation Mechanism of This Bifunctional Enzyme.
J.Biol.Chem., 282, 2007
|
|
4C3Z
 
 | | Nucleotide-free crystal structure of nucleotide-binding domain 1 from human MRP1 supports a general-base catalysis mechanism for ATP hydrolysis. | | Descriptor: | MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 1, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Magnard, S, Falson, P, Di Pietro, A, Baubichon-Cortay, H. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleotide-Free Crystal Structure of Nucleotide-Binding Domain 1 from Human Abcc1 Supports a 'General-Base Catalysis' Mechanism for ATP Hydrolysis.
Biochem.Pharm., 3, 2014
|
|
2Y5Y
 
 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4D5U
 
 | | Structure of OmpF in I2 | | Descriptor: | OUTER MEMBRANE PROTEIN F | | Authors: | Chaptal, V, Kilburg, A, Flot, D, Wiseman, B, Aghajari, N, Jault, J.M, Falson, P. | | Deposit date: | 2014-11-07 | | Release date: | 2015-12-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Two Different Centered Monoclinic Crystals of the E. Coli Outer-Membrane Protein Ompf Originate from the Same Building Block.
Biochim.Biophys.Acta, 1858, 2016
|
|
2QVK
 
 | |
2QVM
 
 | | The second Ca2+-binding domain of the Na+-Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Chaptal, V, Mercado Besserer, G, Abramson, J, Cascio, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The second Ca2+-binding domain of the Na+ Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7OW8
 
 | | CryoEM structure of the ABC transporter BmrA E504A mutant in complex with ATP-Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
7BG4
 
 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP, Mg, and Rhodamine 6G solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA, ... | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
4JFB
 
 | | Crystal structure of OmpF in C2 with tNCS | | Descriptor: | Outer membrane protein F | | Authors: | Wiseman, B, Kilburg, A, Chaptal, V, Reyes-Meija, G.C, Sarwan, J, Falson, P, Jault, J.M. | | Deposit date: | 2013-02-28 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Stubborn contaminants: influence of detergents on the purity of the multidrug ABC transporter BmrA
PLoS ONE, 9, 2014
|
|
6R81
 
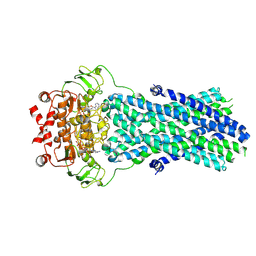 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2019-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
2XQ2
 
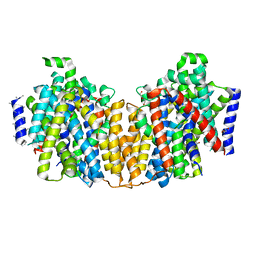 | | Structure of the K294A mutant of vSGLT | | Descriptor: | DI(HYDROXYETHYL)ETHER, SODIUM/GLUCOSE COTRANSPORTER | | Authors: | Watanabe, A, Choe, S, Chaptal, V, Rosenberg, J.M, Wright, E.M, Grabe, M, Abramson, J. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Mechanism of Sodium and Substrate Release from the Binding Pocket of Vsglt
Nature, 468, 2010
|
|
8QQ7
 
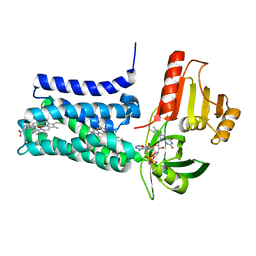 | | Structure of SpNOX: a Bacterial NADPH oxidase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-binding FR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Chaptal, V, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
8SUE
 
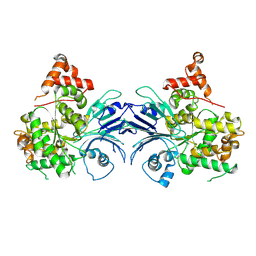 | | Human asparagine synthetase (apo-ASNS) | | Descriptor: | Asparagine synthetase [glutamine-hydrolyzing] | | Authors: | Coricello, A, Zhu, W, Lupia, A, Gratteri, C, Vos, M, Chaptal, V, Alcaro, S, Takagi, Y, Richards, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Human asparagine synthetase (apo-ASNS)
To Be Published
|
|
