4M4C
 
 | | Crystal structure of Rhodostomin ARGDP mutant | | Descriptor: | SULFATE ION, Zinc metalloproteinase/disintegrin | | Authors: | Chang, Y.T, Jeng, W.Y, Shiu, J.H, Chen, C.Y, Chuang, W.J. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of C-terminal proline residue adjacent to the RGD motif in rhodostomin on its activity and structure
To be Published
|
|
7YPL
 
 | |
6LSQ
 
 | |
7X4S
 
 | |
7X4V
 
 | |
7X4Z
 
 | |
7X44
 
 | |
7X43
 
 | |
7X4D
 
 | |
7X41
 
 | |
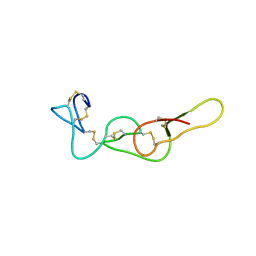
4R5U
 
 | |
4R5R
 
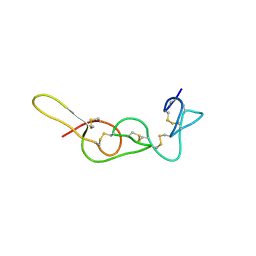 | |
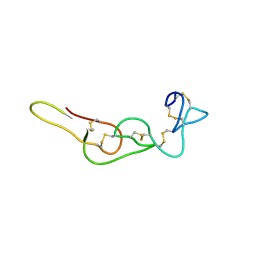
4RQG
 
 | |
3UCI
 
 | | Crystal structure of Rhodostomin ARLDDL mutant | | Descriptor: | disintegrin | | Authors: | Shiu, J.H, Chen, C.Y, Chen, Y.C, Chang, Y.T, Chang, Y.S, Huang, C.H, Chuang, W.J. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
2PJF
 
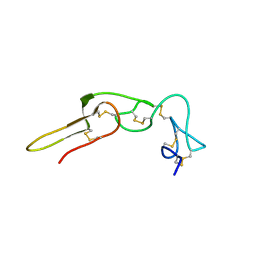 | | Solution structure of rhodostomin | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chang, Y.T. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
