2BJF
 
 | | Crystal Structure of Conjugated Bile Acid Hydrolase from Clostridium perfringens in Complex with Reaction Products Taurine and Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 2-AMINOETHANESULFONIC ACID, CHOLOYLGLYCINE HYDROLASE, ... | | Authors: | Rossocha, M, Schultz-Heienbrok, R, Von Moeller, H, Coleman, J.P, Saenger, W. | | Deposit date: | 2005-02-02 | | Release date: | 2005-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Biochemistry, 44, 2005
|
|
2J9M
 
 | | Crystal Structure of CDK2 in complex with Macrocyclic Aminopyrimidine | | Descriptor: | 6-BROMO-13-THIA-2,4,8,12,19-PENTAAZATRICYCLO[12.3.1.1~3,7~]NONADECA-1(18),3(19),4,6,14,16-HEXAENE 13,13-DIOXIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Luecking, U, Siemeister, G, Briem, H, Krueger, M, Lienau, P, Jautelat, R. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic Aminopyrimidines as Multitarget Cdk and Vegf-R Inhibitors with Potent Antiproliferative Activities.
Chemmedchem, 2, 2007
|
|
2LRE
 
 | | The solution structure of the dimeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2LD0
 
 | |
2LD2
 
 | |
3JWP
 
 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
7N2U
 
 | | Elongating 70S ribosome complex in a hybrid-H1 pre-translocation (PRE-H1) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N30
 
 | | Elongating 70S ribosome complex in a hybrid-H2* pre-translocation (PRE-H2*) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-30 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N1P
 
 | | Elongating 70S ribosome complex in a classical pre-translocation (PRE-C) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, S.K, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2V
 
 | | Elongating 70S ribosome complex in a spectinomycin-stalled intermediate state of translocation bound to EF-G in an active, GTP conformation (INT1) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2C
 
 | | Elongating 70S ribosome complex in a fusidic acid-stalled intermediate state of translocation bound to EF-G(GDP) (INT2) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N31
 
 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
4UV3
 
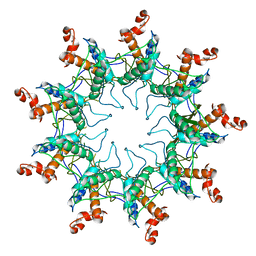 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
3J1Q
 
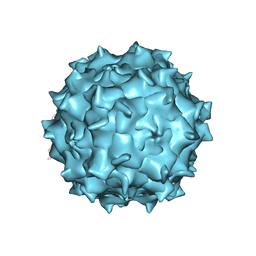 | | Structure of AAV-DJ, a Retargeted Gene Therapy Vector: Cryo-Electron Microscopy at 4.5A resolution | | Descriptor: | Adeno-associated virus DJ | | Authors: | Lerch, T.F, O'Donnell, J.K, Meyer, N.L, Xie, Q, Taylor, K.A, Stagg, S.M, Chapman, M.S. | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of AAV-DJ, a retargeted gene therapy vector: cryo-electron microscopy at 4.5 A resolution.
Structure, 20, 2012
|
|
2PDA
 
 | |
6XWL
 
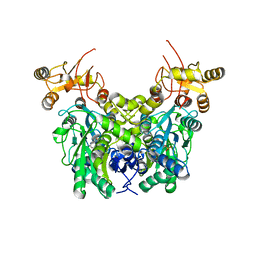 | | Cystathionine beta-synthase from Toxoplasma gondii | | Descriptor: | Cystathionine beta-synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Fernandez-Rodriguez, C, Oyenarte, I, Conter, C, Gonzalez-Recio, I, Quintana, I, Martinez-Chantar, M, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2020-01-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural insight into the unique conformation of cystathionine beta-synthase from Toxoplasma gondii .
Comput Struct Biotechnol J, 19, 2021
|
|
6OEA
 
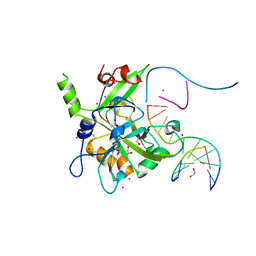 | | Crystal structure of HMCES SRAP domain in complex with longer 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*TP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2PBF
 
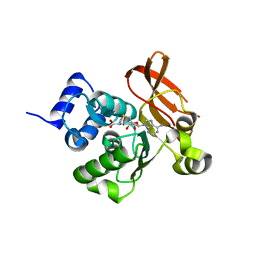 | | Crystal structure of a putative protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase (PCMT) from Plasmodium falciparum in complex with S-adenosyl-L-homocysteine | | Descriptor: | Protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wernimont, A.K, Hassanali, A, Lin, L, Lew, J, Zhao, Y, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase (PCMT) from Plasmodium falciparum in complex with S-adenosyl-L-homocysteine
To be Published
|
|
2P7Q
 
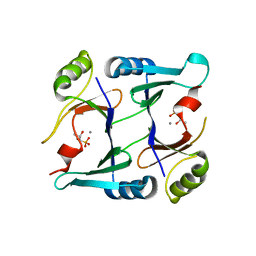 | | Crystal structure of E126Q mutant of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and 1S,2S-dihydroxypropylphosphonic acid | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, [(1S,2S)-1,2-DIHYDROXYPROPYL]PHOSPHONIC ACID | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
5IG3
 
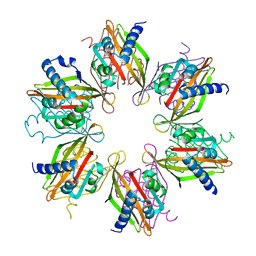 | | Crystal structure of the human CaMKII-alpha hub | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | McSpadden, E, Cao, Y.M, Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
2P7L
 
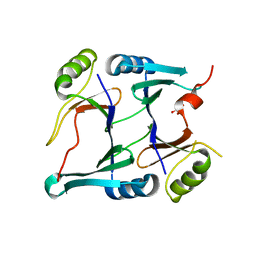 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 5.75 | | Descriptor: | GLYCEROL, Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7P
 
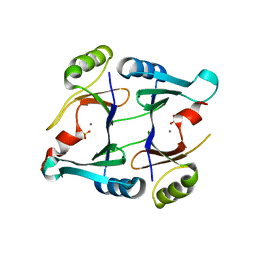 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and sulfate ion | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, SULFATE ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7O
 
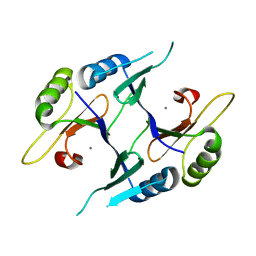 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes (tetragonal form) | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7K
 
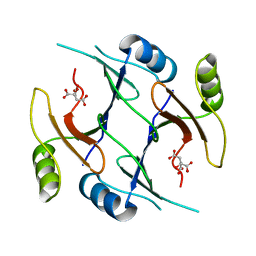 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes (hexagonal form) | | Descriptor: | CITRIC ACID, Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
