6Q5T
 
 | |
6G3N
 
 | |
6S2Q
 
 | |
6S2R
 
 | |
9GZJ
 
 | |
9GZK
 
 | |
7AGY
 
 | |
7AHT
 
 | |
7AGV
 
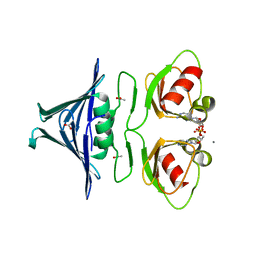 | | High-resolution structure of the K+/H+ antiporter subunit KhtT in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETATE ION, CALCIUM ION, ... | | Authors: | Cereija, T.B, Guerra, J.P, Morais-Cabral, J.H. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | c-di-AMP, a likely master regulator of bacterial K + homeostasis machinery, activates a K + exporter.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AGW
 
 | |
7AHM
 
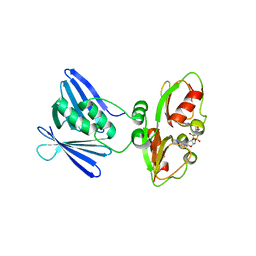 | | Low-resolution structure of the K+/H+ antiporter subunit KhtT in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, K(+)/H(+) antiporter subunit KhtT | | Authors: | Cereija, T.B, Guerra, J.P, Morais-Cabral, J.H. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | c-di-AMP, a likely master regulator of bacterial K + homeostasis machinery, activates a K + exporter.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8BZ7
 
 | |
8BZ4
 
 | |
8BZ6
 
 | |
8BZ8
 
 | |
8BZ5
 
 | |
5OHZ
 
 | |
5OI1
 
 | |
5OHC
 
 | |
5OIW
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycerate | | Descriptor: | (2R)-2-(alpha-D-glucopyranosyloxy)-3-hydroxypropanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5OI0
 
 | |
5OJ4
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with mannosylglycerate | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5OJU
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with glucosylglycerate | | Descriptor: | (2R)-2-(alpha-D-glucopyranosyloxy)-3-hydroxypropanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-24 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5OIE
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with serine and glycerol | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SERINE, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
5OIV
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D43A variant in complex with serine and glycerol | | Descriptor: | GLYCEROL, GLYCINE, Hydrolase, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
