1YZ1
 
 | |
3PY7
 
 | |
2OQB
 
 | |
1ASZ
 
 | | THE ACTIVE SITE OF YEAST ASPARTYL-TRNA SYNTHETASE: STRUCTURAL AND FUNCTIONAL ASPECTS OF THE AMINOACYLATION REACTION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTYL-tRNA SYNTHETASE, T-RNA (75-MER) | | Authors: | Cavarelli, J, Rees, B, Thierry, J.C, Moras, D. | | Deposit date: | 1995-01-19 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction.
EMBO J., 13, 1994
|
|
1BS2
 
 | | YEAST ARGINYL-TRNA SYNTHETASE | | Descriptor: | ARGININE, PROTEIN (ARGINYL-TRNA SYNTHETASE) | | Authors: | Cavarelli, J, Delagouute, B, Eriani, G, Gangloff, J, Moras, D. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | L-arginine recognition by yeast arginyl-tRNA synthetase.
EMBO J., 17, 1998
|
|
1AGJ
 
 | | EPIDERMOLYTIC TOXIN A FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | EPIDERMOLYTIC TOXIN A | | Authors: | Cavarelli, J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Staphylococcus aureus epidermolytic toxin A, an atypic serine protease, at 1.7 A resolution.
Structure, 5, 1997
|
|
3B3J
 
 | | The 2.55 A crystal structure of the apo catalytic domain of coactivator-associated arginine methyl transferase I(CARM1:28-507, residues 28-146 and 479-507 not ordered) | | Descriptor: | BENZAMIDINE, Histone-arginine methyltransferase CARM1 | | Authors: | Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, Cavarelli, J. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.
Embo J., 26, 2007
|
|
2QEN
 
 | | The walker-type atpase paby2304 of pyrococcus abyssi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, WALKER-TYPE ATPASE | | Authors: | Uhring, M, Bey, G, Lecompte, O, Moras, D, Poch, O, Cavarelli, J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The walker-type atpase paby2304 of pyrococcus abyssi
To be Published
|
|
7QRD
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_10-25 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
7QPH
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_22-31 K27 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone H3 22-31 K27 acetylated, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-04 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
8PJD
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 94-445 | | Descriptor: | ACETATE ION, Protein arginine N-methyltransferase 2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 94-445
To Be Published
|
|
6TQR
 
 | | The crystal structure of the MSP domain of human VAP-A in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | CHLORIDE ION, StAR-related lipid transfer protein 3, Vesicle-associated membrane protein-associated protein A | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
3O0A
 
 | | Crystal structure of the wild type CP1 hydrolitic domain from Aquifex Aeolicus leucyl-trna | | Descriptor: | Leucyl-tRNA synthetase subunit alpha | | Authors: | Cura, V, Olieric, N, Wang, E.-D, Moras, D, Eriani, G, Cavarelli, J. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the Wild Type and Two Mutants of the Cp1 Hydrolytic Domain from Aquifex Aeolicus Leucyl-tRNA Synthetase
To be Published
|
|
6TQT
 
 | | The crystal structure of the MSP domain of human MOSPD2. | | Descriptor: | 1,2-ETHANEDIOL, Motile sperm domain-containing protein 2, PHOSPHATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
4X33
 
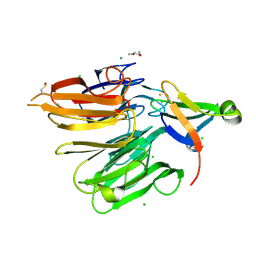 | | Structure of the Elongator cofactor complex Kti11/Kti13 at 1.45A | | Descriptor: | 1,2-DIMETHOXYETHANE, CHLORIDE ION, Diphthamide biosynthesis protein 3, ... | | Authors: | Kolaj-Robin, O, McEwen, A.G, Cavarelli, J, Seraphin, B. | | Deposit date: | 2014-11-27 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification.
Febs J., 282, 2015
|
|
8BVA
 
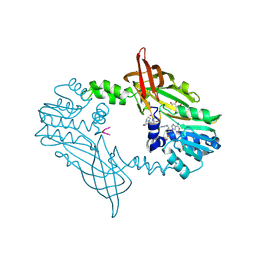 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-02 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126
To Be Published
|
|
8C1J
 
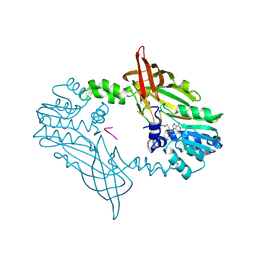 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Protein arginine N-methyltransferase 2, RNA-binding protein Rsf1-like | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30
To Be Published
|
|
1EOV
 
 | | FREE ASPARTYL-TRNA SYNTHETASE (ASPRS) (E.C. 6.1.1.12) FROM YEAST | | Descriptor: | ASPARTYL-TRNA SYNTHETASE | | Authors: | Sauter, C, Lorber, B, Cavarelli, J, Moras, D, Giege, R. | | Deposit date: | 2000-03-24 | | Release date: | 2000-09-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free yeast aspartyl-tRNA synthetase differs from the tRNA(Asp)-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain.
J.Mol.Biol., 299, 2000
|
|
7NUD
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML734 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[3-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]propylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7NUE
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
4Z9V
 
 | | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL | | Descriptor: | BICARBONATE ION, Bcl-2-like protein 1,APOPTOSIS REGULATOR BCL-XL, SODIUM ION, ... | | Authors: | Cura, V, Agez, M, Thebault, S, Cavarelli, J. | | Deposit date: | 2015-04-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL.
Sci Rep, 6, 2016
|
|
1JMM
 
 | | Crystal structure of the V-region of Streptococcus mutans antigen I/II | | Descriptor: | SODIUM ION, protein I/II V-region | | Authors: | Troffer-Charlier, N, Ogier, J, Moras, D, Cavarelli, J. | | Deposit date: | 2001-07-19 | | Release date: | 2002-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the V-region of Streptococcus mutans Antigen I/II at 2.4 a Resolution Suggests a Sugar Preformed Binding Site
J.Mol.Biol., 318, 2002
|
|
4GIZ
 
 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
1F7U
 
 | | CRYSTAL STRUCTURE OF THE ARGINYL-TRNA SYNTHETASE COMPLEXED WITH THE TRNA(ARG) AND L-ARG | | Descriptor: | ARGININE, ARGINYL-TRNA SYNTHETASE, SULFATE ION, ... | | Authors: | Delagoutte, B, Moras, D, Cavarelli, J. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding
EMBO J., 19, 2000
|
|
1F7V
 
 | |
