6SW8
 
 | | Crystal structure of the NS1 (H7N1) RNA-binding domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural protein 1 | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
6SX0
 
 | | Specific dsRNA recognition by wild type H7N1 NS1 RNA-binding domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Non-structural protein 1, ... | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
6SX2
 
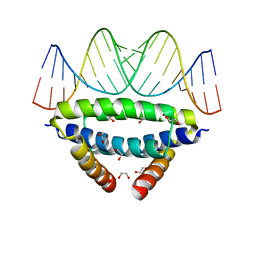 | | dsRNA recognition by R38AK41A mutant of H7N1 NS1 RNA Binding Domain | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 1, RNA (5'-R(*GP*GP*UP*AP*AP*CP*UP*GP*UP*UP*AP*CP*AP*GP*UP*UP*AP*CP*C)-3') | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
6RO2
 
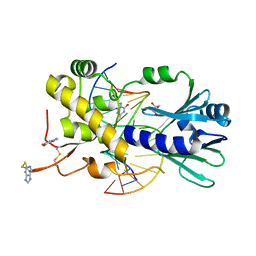 | | The crystal structure of a complex between the LlFpg protein, a THF-DNA and an inhibitor | | Descriptor: | 2-sulfanyl-1,9-dihydro-6H-purin-6-one, DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Coste, F, Goffinont, S, Castaing, B. | | Deposit date: | 2019-05-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights.
Int J Mol Sci, 21, 2020
|
|
6RP0
 
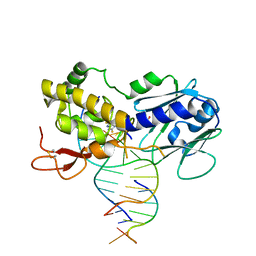 | | The crystal structure of a complex between the LlFpg protein, a THF-DNA and an inhibitor | | Descriptor: | 2,8-dithioxo-1,2,3,7,8,9-hexahydro-6H-purin-6-one, DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Coste, F, Goffinont, S, Castaing, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights.
Int J Mol Sci, 21, 2020
|
|
7OLB
 
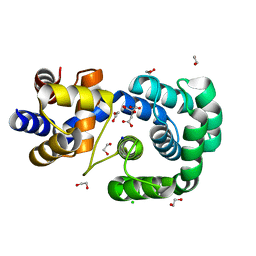 | | Crystal structure of Pab-AGOG, an 8-oxoguanine DNA glycosylase from Pyrococcus abyssi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OUE
 
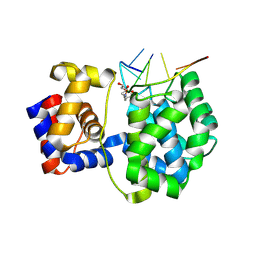 | | Crystal structure of a trapped Pab-AGOG/single-standed DNA covalent intermediate | | Descriptor: | DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), N-glycosylase/DNA lyase, PHOSPHATE ION, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OU3
 
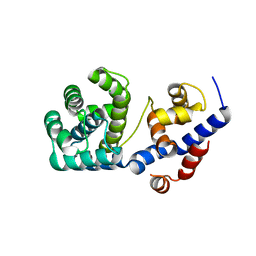 | | Crystal structure of Tga-AGOG, an 8-oxoguanine DNA glycosylase from Thermococcus gammatolerans | | Descriptor: | CHLORIDE ION, GLYCEROL, N-glycosylase/DNA lyase | | Authors: | Coste, F, Confalonieri, F, Castaing, B. | | Deposit date: | 2021-06-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P8L
 
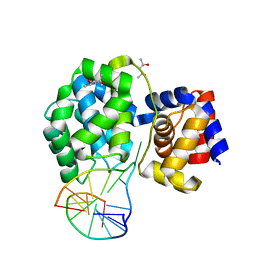 | | Crystal structure of Pyrococcus abyssi 8-oxoguanine DNA glycosylase (PabAGOG) in complex with dsDNA containing cytosine opposite to 8-oxoG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(8OG)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Flament, D, Castaing, B. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P0W
 
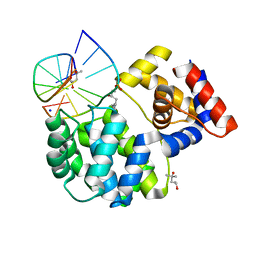 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing thymine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OY7
 
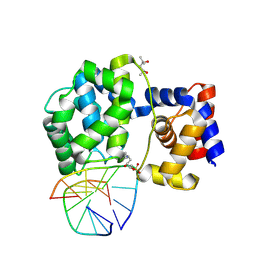 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing cytosine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P9Z
 
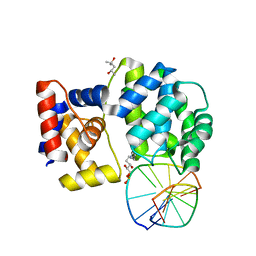 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing adenine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OLI
 
 | | Crystal structure of Pab-AGOG in complex with 8-oxoguanosine | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-8-OXOGUANOSINE, N-glycosylase/DNA lyase | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
