8VO1
 
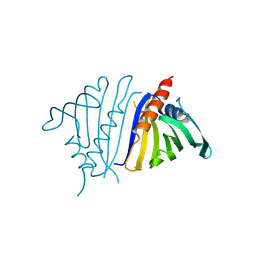 | | Pathogenesis related 10-10 C155S mutant | | Descriptor: | Pathogenesis related 10-10 C155S mutant | | Authors: | Carr, S.C, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a ligand-triggered intermolecular disulfide switch in a major latex protein from opium poppy.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6FPI
 
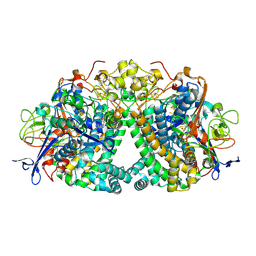 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
8VO2
 
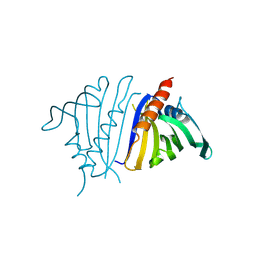 | | Pathogenesis related 10-10 C59S mutant | | Descriptor: | Pathogenesis related 10-10 C59S mutant | | Authors: | Carr, S.C, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of a ligand-triggered intermolecular disulfide switch in a major latex protein from opium poppy.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8VO3
 
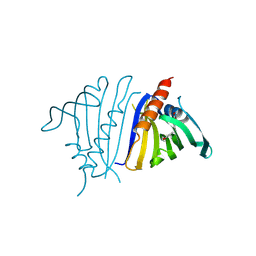 | | Pathogenesis related 10-10 C59S mutant papaverine complex | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, Pathogenesis related 10-10 C59S mutant | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of a ligand-triggered intermolecular disulfide switch in a major latex protein from opium poppy.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7MBF
 
 | | codeinone reductase isoform 1.3 Apo form | | Descriptor: | NADPH-dependent codeinone reductase 1-3 | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of codeinone reductase reveal novel insights into aldo-keto reductase function in benzylisoquinoline alkaloid biosynthesis.
J.Biol.Chem., 297, 2021
|
|
7NEM
 
 | | Hydrogenase-2 variant R479K - anaerobically oxidised form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B. | | Deposit date: | 2021-02-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A comprehensive structural and kinetic investigation of the role of the active-site argininein bidirectional hydrogen activation by the [NiFe]-hydrogenase "Hyd-2) from Escherichia coli
To Be Published
|
|
6GAN
 
 | | Structure of fully reduced Hydrogenase (Hyd-2) variant E14Q | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GAM
 
 | | Structure of E14Q variant of E. coli hydrogenase-2 (as-isolated enzyme) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6G7M
 
 | | Four-site variant (Y222C, C197S, C432S, C433S) of E. coli hydrogenase-2 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L, Beaton, S.E. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Direct visible light activation of a surface cysteine-engineered [NiFe]-hydrogenase by silver nanoclusters
Energy Environ Sci, 2019
|
|
6G7R
 
 | | Structure of fully reduced variant E28Q of E. coli hydrogenase-1 at pH 8 | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
5LRY
 
 | | E coli [NiFe] Hydrogenase Hyd-1 mutant E28D | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J.Am.Chem.Soc., 140, 2018
|
|
6GAL
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q collected at pH 10 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6FPW
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6FPO
 
 | | High resolution structure of native Hydrogenase (Hyd-1) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
5LMM
 
 | | Structure of E coli Hydrogenase Hyd-1 mutant E28Q | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Kinetic consequences of re-engineering the outer shell "canopy" above the active site of a [NiFe]-hydrogenase.
To Be Published
|
|
5JRD
 
 | | E. coli Hydrogenase-1 variant P508A | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Armstrong, F.A, Evans, R.M, Brooke, E.J, Islam, S.T.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Biochemistry, 56, 2017
|
|
7UQN
 
 | | Pathogenesis related 10-10 noscapine complex | | Descriptor: | Pathogenesis Related 10-10 protein, noscapine | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
7UQL
 
 | | Pathogenesis related 10-10 app from | | Descriptor: | Pathogenesis Related 10-10 protein | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
7UQM
 
 | | Pathogenesis related 10-10 papaverine complex | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, Pathogenesis Related 10-10 protein | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
6EHS
 
 | | E. coli Hydrogenase-2 chemically reduced structure | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, DITHIONITE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Evans, R.M, Beaton, S.E, Armstrong, F.A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6EN9
 
 | | E. coli Hydrogenase-2 (hydrogen reduced form) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
9H8I
 
 | | Crystallization of B. licheniformis levanase | | Descriptor: | 1,2-ETHANEDIOL, BICINE, CALCIUM ION, ... | | Authors: | Carr, S, Cruz-Migoni, A, Porras-Dominguez, J.R, Van den Ende, W, Lopez-Munguia Canales, A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Understanding the Endo- and Exo-mechanisms Involved in the Enzymatic Hydrolysis of Levan and Inulin Polymers.
J.Agric.Food Chem., 73, 2025
|
|
6EHQ
 
 | | E. coli Hydrogenase-2 (as isolated form). | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
7UQO
 
 | | Pathogenesis related 10-10 (S)-tetrahydropapaverine complex | | Descriptor: | (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pathogenesis Related 10-10 protein | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
1E44
 
 | | ribonuclease domain of colicin E3 in complex with its immunity protein | | Descriptor: | 1,2-ETHANEDIOL, COLICIN E3, IMMUNITY PROTEIN | | Authors: | Carr, S, Walker, D, James, R, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of a Ribosome Inactivating Ribonuclease: The Crystal Structure of the Cytotoxic Domain of Colicin E3 in Complex with its Immunity Protein
Structure, 8, 2000
|
|
