4LFB
 
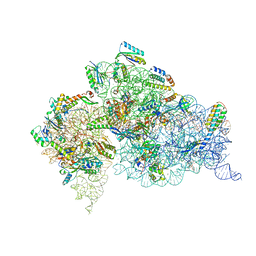 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, NEOMYCIN, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
to be published, 2013
|
|
4LFC
 
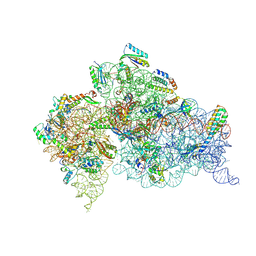 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: |
to be published, 2013
|
|
4LF9
 
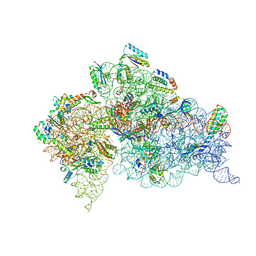 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDROXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, MAGNESIUM ION, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.2811 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
to be published, 2013
|
|
4LF7
 
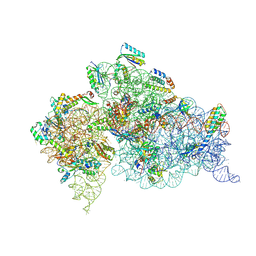 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, PAROMOMYCIN, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.1484 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
To be Published, 2013
|
|
4V0I
 
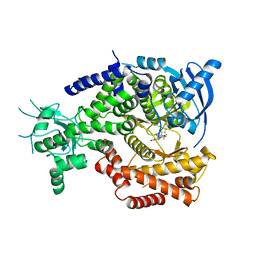 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
6QXM
 
 | | Cryo-EM structure of T7 bacteriophage portal protein, 12mer, open valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6R21
 
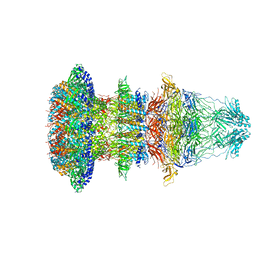 | | Cryo-EM structure of T7 bacteriophage fiberless tail complex | | Descriptor: | Portal protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Cuervo, A, Fabrega-Ferrer, M, Machon, C, Conesa, J.J, Perez-Ruiz, M, Coll, M, Carrascosa, J.L. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6YI9
 
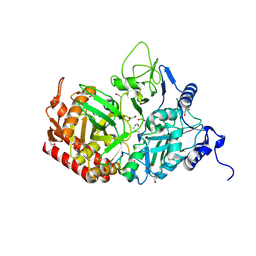 | | Crystal structure of the rat cytosolic PCK1, acetylated on Lys244 | | Descriptor: | 1,2-ETHANEDIOL, Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | | Authors: | Latorre-Muro, P, Baeza, J, Hurtado-Guerrero, R, Hicks, T, Delso, I, Hernandez-Ruiz, C, Velazquez-Campoy, A, Lawton, A.J, Angulo, J, Denu, J.M, Carrodeguas, J.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Self-acetylation at the active site of phosphoenolpyruvate carboxykinase (PCK1) controls enzyme activity.
J.Biol.Chem., 296, 2021
|
|
6QWP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6YSZ
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core protein intermediate assembly | | Descriptor: | Internal virion protein gp15 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YT5
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15-gp16 core complex intermediate assembly | | Descriptor: | Internal virion protein gp15, Peptidoglycan transglycosylase gp16 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6QX5
 
 | | Crystal structure of T7 bacteriophage portal protein, 12mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6RVR
 
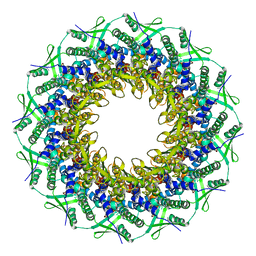 | | Atomic structure of the Epstein-Barr portal, structure I | | Descriptor: | Portal protein | | Authors: | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
5ND1
 
 | | Viral evolution results in multiple, surface-allocated enzymatic activities in a fungal double-stranded RNA virus | | Descriptor: | Capsid protein | | Authors: | Mata, C.P, Luque, D, Gomez Blanco, J, Rodriguez, J.M, Suzuki, N, Ghabrial, S.A, Carrascosa, J.L, Trus, B.L, Caston, J.R. | | Deposit date: | 2017-03-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses.
PLoS Pathog., 13, 2017
|
|
3NET
 
 | |
3IZG
 
 | | Bacteriophage T7 prohead shell EM-derived atomic model | | Descriptor: | Major capsid protein 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Agirrezabala, X, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
3J4A
 
 | | Structure of gp8 connector protein | | Descriptor: | Head-to-tail joining protein | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
3J4B
 
 | | Structure of T7 gatekeeper protein (gp11) | | Descriptor: | Tail tubular protein A | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
3J3I
 
 | | Penicillium chrysogenum virus (PcV) capsid structure | | Descriptor: | Capsid protein | | Authors: | Luque, D, Gomez-Blanco, J, Garriga, D, Brilot, A, Gonzalez, J.M, Havens, W.H, Carrascosa, J.L, Trus, B.L, Verdaguer, N, Grigorieff, N, Ghabrial, S.A, Caston, J.R. | | Deposit date: | 2013-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3O66
 
 | | Crystal structure of glycine betaine/carnitine/choline ABC transporter | | Descriptor: | ACETATE ION, Glycine betaine/carnitine/choline ABC transporter, TRIETHYLENE GLYCOL | | Authors: | Chang, C, Bigelow, L, Carroll, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of glycine betaine/carnitine/choline ABC transporter
To be Published
|
|
3ZUE
 
 | | Rabbit Hemorrhagic Disease Virus (RHDV)capsid protein | | Descriptor: | CAPSID STRUCTURAL PROTEIN VP60 | | Authors: | Luque, D, Gonzalez, J.M, Gomez-Blanco, J, Marabini, R, Chichon, J, Mena, I, Angulo, I, Carrascosa, J.L, Verdaguer, N, Trus, B.L, Barcena, J, Caston, J.R. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Epitope Insertion at the N-Terminal Molecular Switch of the Rabbit Hemorrhagic Disease Virus T=3 Capsid Protein Leads to Larger T=4 Capsids.
J.Virol., 86, 2012
|
|
6TJP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve - P212121 | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Fernandez, F.J, Machon, C, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Using a partial atomic model from medium-resolution cryo-EM to solve a large crystal structure.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2XVR
 
 | | Phage T7 empty mature head shell | | Descriptor: | MAJOR CAPSID PROTEIN 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Molecular Rearrangements Involved in the Capsid Shell Maturation of Bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
6RVS
 
 | | Atomic structure of the Epstein-Barr portal, structure II | | Descriptor: | Portal protein | | Authors: | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
2YFQ
 
 | | Crystal structure of Glutamate dehydrogenase from Peptoniphilus asaccharolyticus | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Oliveira, T, Panjikar, S, Carrigan, J.B, Sharkey, M.A, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-07 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of Nad-Dependent Peptoniphilus Asaccharolyticus Glutamate Dehydrogenase Reveals Determinants of Cofactor Specificity.
J.Struct.Biol., 177, 2012
|
|
