6PO4
 
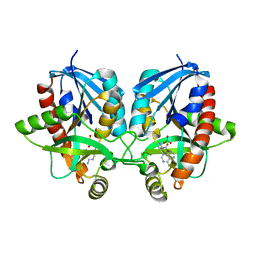 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
5T1V
 
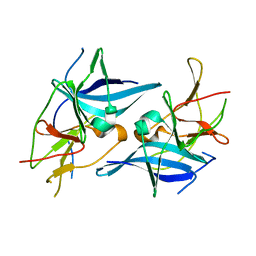 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | Descriptor: | NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
6E5Y
 
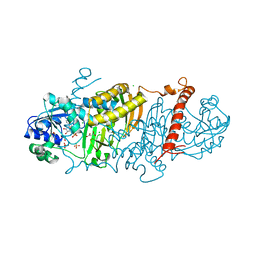 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
6N0I
 
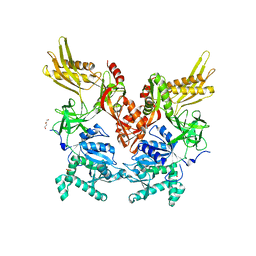 | | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongation factor G 2, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida.
To Be Published
|
|
6N7F
 
 | | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD. | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Shabalin, I.G, Grabowski, M, Olphie, A, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.
To Be Published
|
|
6BK7
 
 | | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis | | Descriptor: | Elongation factor G, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis.
To be Published
|
|
6CN1
 
 | | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
6B4O
 
 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | Authors: | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
5TY0
 
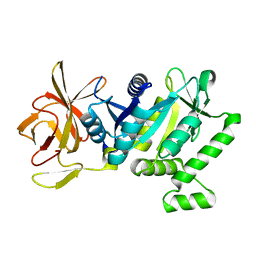 | | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila. | | Descriptor: | Elongation factor G, SODIUM ION, beta-D-glucopyranose | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila.
To Be Published
|
|
5VDN
 
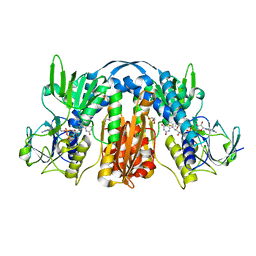 | | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Glutathione oxidoreductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD.
To Be Published
|
|
5U47
 
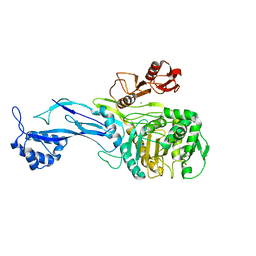 | | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, Penicillin binding protein 2X | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-03 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus.
To Be Published
|
|
5U1O
 
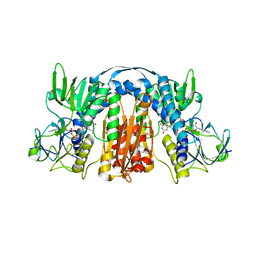 | | 2.3 Angstrom Resolution Crystal Structure of Glutathione Reductase from Vibrio parahaemolyticus in Complex with FAD. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3 Angstrom Resolution Crystal Structure of Glutathione Reductase from Vibrio parahaemolyticus in Complex with FAD.
To Be Published
|
|
5TV2
 
 | | Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6 | | Descriptor: | Elongation factor G | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6
To Be Published
|
|
5TU0
 
 | | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose | | Descriptor: | Lmo2125 protein, TARTRONATE, TRIETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose.
To Be Published
|
|
5TXG
 
 | | Crystal structure of the Zika virus NS3 helicase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NS3 helicase, POTASSIUM ION | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the Zika virus NS3 helicase.
To be published
|
|
5UPR
 
 | | X-ray structure of a putative triosephosphate isomerase from Toxoplasma gondii ME49 | | Descriptor: | CHLORIDE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Cardona-Correa, A, Bishop, B, Anderson, W.F, Ngo, H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a putative triosephosphate isomerase from Toxoplasma gondii ME49
To Be Published
|
|
5KZS
 
 | | Listeria monocytogenes internalin-like protein lmo2027 | | Descriptor: | Putative cell surface protein, similar to internalin proteins | | Authors: | Light, S.H, Nocadello, S, Minasov, G, Cardona-Correa, A, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
