5WCR
 
 | | Phosphotriesterase variant R0deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R0, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-01 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphotriesterase variant R0deltaL7
To Be Published
|
|
5WCP
 
 | | Phosphotriesterase variant S7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-01 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphotriesterase variant S7
To Be Published
|
|
5WIZ
 
 | | Phosphotriesterase variant S5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-21 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Phosphotriesterase variant S5
To Be Published
|
|
6BH7
 
 | | Phosphotriesterase variant R18+254S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18+254S
To Be Published
|
|
6BHL
 
 | | Phosphotriesterase variant S5deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant S5deltaL7
To Be Published
|
|
6DCF
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6BZO
 
 | | Mtb RNAP Holo/RbpA/Fidaxomicin/upstream fork DNA | | Descriptor: | DNA (26-MER), DNA (32-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2017-12-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
1HQM
 
 | | CRYSTAL STRUCTURE OF THERMUS AQUATICUS CORE RNA POLYMERASE-INCLUDES COMPLETE STRUCTURE WITH SIDE-CHAINS (EXCEPT FOR DISORDERED REGIONS)-FURTHER REFINED FROM ORIGINAL DEPOSITION-CONTAINS ADDITIONAL SEQUENCE INFORMATION | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Minakhin, L, Bhagat, S, Brunning, A, Campbell, E.A, Darst, S.A, Ebright, R.H, Severinov, K. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6CCE
 
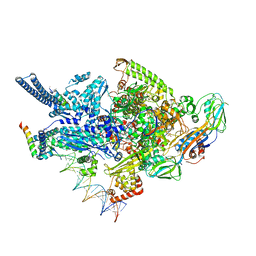 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (57-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6CCV
 
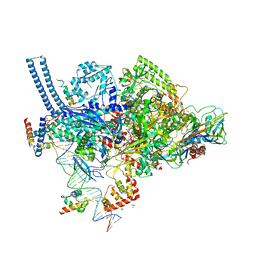 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Rifampicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6EEC
 
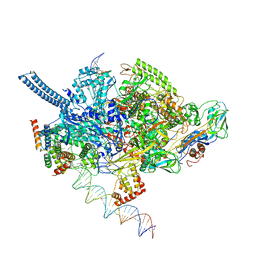 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter captured by Corallopyronin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
1L9Z
 
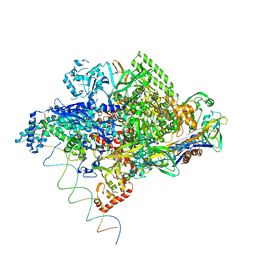 | | Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Campbell, E.A, Muzzin, O, Darst, S.A. | | Deposit date: | 2002-03-27 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.
Science, 296, 2002
|
|
1OTT
 
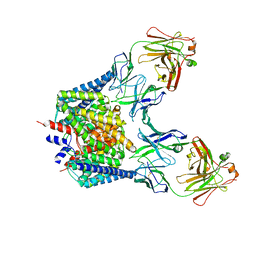 | | Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1TH8
 
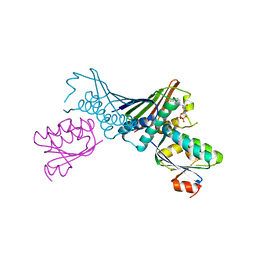 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigian, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1OTS
 
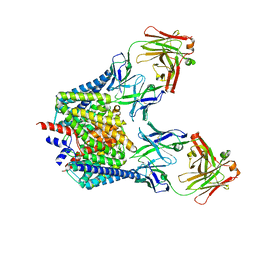 | | Structure of the Escherichia coli ClC Chloride channel and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (heavy chain), Fab fragment (light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-22 | | Release date: | 2003-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OTU
 
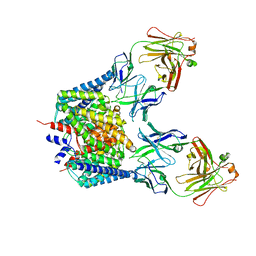 | | Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1TIL
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA:Poised for phosphorylation complex with ATP, crystal form II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1THN
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1TID
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: Poised for phosphorylation complex with ATP, crystal form I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
