1WND
 
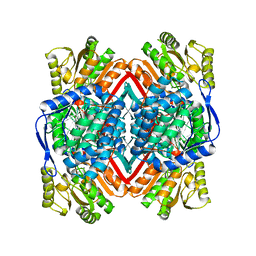 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase as determined by kinetics and crystal structure | | Descriptor: | CALCIUM ION, Putative betaine aldehyde dehydrogenase | | Authors: | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
1QZ8
 
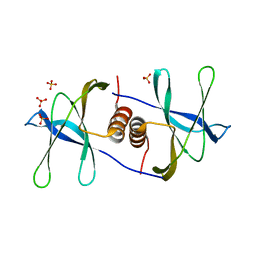 | | Crystal structure of SARS coronavirus NSP9 | | Descriptor: | SULFATE ION, polyprotein 1ab | | Authors: | Egloff, M.P, Ferron, F, Campanacci, V, Longhi, S, Rancurel, C, Dutartre, H, Snijder, E.J, Gorbalenya, A.E, Cambillau, C, Canard, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1GPL
 
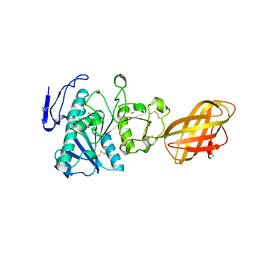 | | RP2 LIPASE | | Descriptor: | CALCIUM ION, RP2 LIPASE | | Authors: | Withers-Martinez, C, Cambillau, C. | | Deposit date: | 1996-07-13 | | Release date: | 1997-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A pancreatic lipase with a phospholipase A1 activity: crystal structure of a chimeric pancreatic lipase-related protein 2 from guinea pig.
Structure, 4, 1996
|
|
1HPL
 
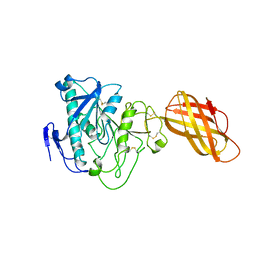 | |
4PSE
 
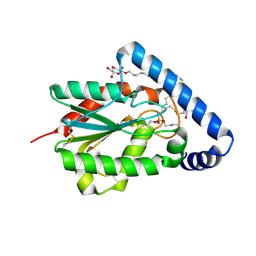 | | Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor | | Descriptor: | Carbohydrate esterase family 5, UNDECYL-PHOSPHINIC ACID BUTYL ESTER, octyl beta-D-glucopyranoside | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
3R72
 
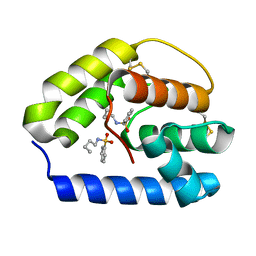 | | Apis mellifera odorant binding protein 5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Odorant binding protein ASP5 | | Authors: | Spinelli, S, Iovinella, I, Lagarde, A, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera odorant binding protein 5
To be Published
|
|
1W8G
 
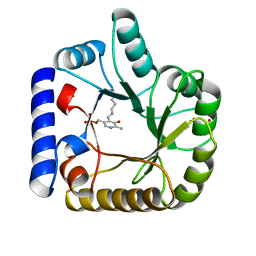 | | CRYSTAL STRUCTURE OF E. COLI K-12 YGGS | | Descriptor: | HYPOTHETICAL UPF0001 PROTEIN YGGS, ISOCITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sulzenbacher, G, Gruez, A, Spinelli, S, Roig-Zamboni, V, Pagot, F, Bignon, C, Vincentelli, R, Cambillau, C. | | Deposit date: | 2004-09-21 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E. Coli K-12 Yggs
To be Published
|
|
1G85
 
 | | CRYSTAL STRUCTURE OF BOVINE ODORANT BINDING PROTEIN COMPLEXED WITH IS NATURAL LIGAND | | Descriptor: | (3R)-oct-1-en-3-ol, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1LOG
 
 | |
1LOB
 
 | |
1LOA
 
 | |
4PSC
 
 | | Structure of cutinase from Trichoderma reesei in its native form. | | Descriptor: | Carbohydrate esterase family 5, GLYCEROL | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
4RGG
 
 | |
1U08
 
 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
1W9A
 
 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
4PSD
 
 | | Structure of Trichoderma reesei cutinase native form. | | Descriptor: | Carbohydrate esterase family 5 | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
1SLB
 
 | | X-RAY CRYSTALLOGRAPHY REVEALS CROSSLINKING OF MAMMALIAN LECTIN (GALECTIN-1) BY BIANTENNARY COMPLEX TYPE SACCHARIDES | | Descriptor: | BOVINE GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-03-12 | | Release date: | 1995-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides.
Nat.Struct.Biol., 1, 1994
|
|
4JEI
 
 | | Nonglycosylated Yarrowia lipolytica LIP2 lipase | | Descriptor: | Lipase 2 | | Authors: | Aloulou, A, Benarouche, A, Puccinelli, D, Spinelli, S, Cavalier, J.-F, Cambillau, C, Carriere, F. | | Deposit date: | 2013-02-27 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of non-glycosylated Yarrowia lipolytica LIP2 lipase
To be published
|
|
1GJQ
 
 | | Pseudomonas aeruginosa cd1 nitrite reductase reduced cyanide complex | | Descriptor: | CYANIDE ION, HEME C, HEME D, ... | | Authors: | Nurizzo, D, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanide Binding to Cd(1) Nitrite Reductase from Pseudomonas Aeruginosa: Role of the Active-Site His369 in Ligand Stabilization.
Biochem.Biophys.Res.Commun., 291, 2002
|
|
1LGB
 
 | |
1HN2
 
 | | CRYSTAL STRUCTURE OF BOVINE OBP COMPLEXED WITH AMINOANTHRACENE | | Descriptor: | (3R)-oct-1-en-3-ol, ANTHRACEN-1-YLAMINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1SLC
 
 | | X-RAY CRYSTALLOGRAPHY REVEALS CROSSLINKING OF MAMMALIAN LECTIN (GALECTIN-1) BY BIANTENNARY COMPLEX TYPE SACCHARIDES | | Descriptor: | BOVINE GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-03-12 | | Release date: | 1995-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides.
Nat.Struct.Biol., 1, 1994
|
|
3BFA
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the Queen mandibular pheromone | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
1DZJ
 
 | | Porcine Odorant Binding Protein Complexed with 2-amino-4-butyl-5-propylselenazole | | Descriptor: | 4-butyl-5-propyl-1,3-selenazol-2-amine, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-01 | | Release date: | 2000-12-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1DZK
 
 | | Porcine Odorant Binding Protein Complexed with pyrazine (2-isobutyl-3-metoxypyrazine) | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-01 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
