4PS2
 
 | | Structure of the C-terminal fragment (87-165) of E.coli EAEC TssB molecule | | Descriptor: | CHLORIDE ION, Putative type VI secretion protein, ZINC ION | | Authors: | Douzi, B, Logger, L, Spinelli, S, Blangy, S, Cambillau, C, Cascales, E. | | Deposit date: | 2014-03-06 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the tube-sheath interface within the Type VI secretion system tail
To be Published
|
|
4QLR
 
 | | Llama nanobody n02 raised against EAEC T6SS TssM | | Descriptor: | Llama nanobody n02 VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
4RGA
 
 | | Phage 1358 receptor binding protein in complex with the trisaccharide GlcNAc-Galf-GlcOMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-galactofuranose-(1-6)-methyl alpha-D-glucopyranoside, Phage 1358 receptor binding protein (ORF20) | | Authors: | Spinelli, S, Mccabe, O, Farenc, C, Tremblay, D, Blangy, S, Oscarson, S, Moineau, S, Cambillau, C. | | Deposit date: | 2014-09-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors.
Mol.Microbiol., 96, 2015
|
|
2J6B
 
 | | crystal structure of AFV3-109, a highly conserved protein from crenarchaeal viruses | | Descriptor: | AFV3-109 | | Authors: | Keller, J, Leulliot, N, Cambillau, C, Campanacci, V, Porciero, S, Prangishvili, D, Cortez, D, Quevillon-Cheruel, S, Van Tilbeurgh, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Afv3-109, a Highly Conserved Protein from Crenarchaeal Viruses.
Virol J., 4, 2007
|
|
1GM6
 
 | | 3-D STRUCTURE OF A SALIVARY LIPOCALIN FROM BOAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Spinelli, S, Vincent, F, Pelosi, P, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Boar Salivary Lipocalin. Three-Dimensional X-Ray Structure and Androsterol/Androstenone Docking Simulations.
Eur.J.Biochem., 269, 2002
|
|
1U08
 
 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
1H8S
 
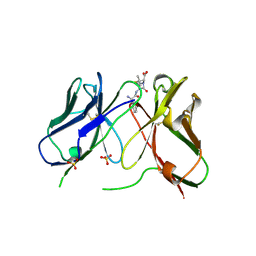 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment complexed with the hapten. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-15 | | Release date: | 2001-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
1H8N
 
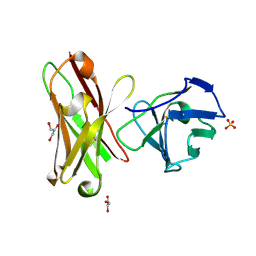 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment from phage-displayed murine antibody libraries | | Descriptor: | GLYCEROL, MUTANT AL2 6E7S9G, SULFATE ION | | Authors: | Jung, S, Spinelli, S, Schimmele, B, Honegger, A, Pugliese, L, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Importance of Framework Residues H6, H7 and H10 in Antibody Heavy Chains: Experimental Evidence for a New Structural Subclassification of Antibody V(H) Domains
J.Mol.Biol., 309, 2001
|
|
1H8O
 
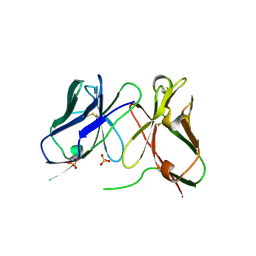 | | Three-dimensional structure of anti-ampicillin single chain Fv fragment. | | Descriptor: | MUTANT AL2 6E7P9G, SULFATE ION | | Authors: | Burmester, J, Spinelli, S, Pugliese, L, Krebber, A, Honegger, A, Jung, S, Schimmele, B, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-14 | | Release date: | 2001-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Selection, Characterization and X-Ray Structure of Anti-Ampicillin Single-Chain Fv Fragments from Phage-Displayed Murine Antibody Libraries
J.Mol.Biol., 309, 2001
|
|
2ACO
 
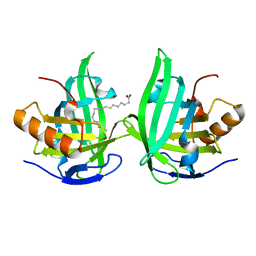 | | Xray structure of Blc dimer in complex with vaccenic acid | | Descriptor: | Outer membrane lipoprotein blc, VACCENIC ACID | | Authors: | Campanacci, V, Bishop, R.E, Reese, L, Blangy, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2005-07-19 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The membrane bound bacterial lipocalin Blc is a functional dimer with binding preference for lysophospholipids.
Febs Lett., 580, 2006
|
|
2H85
 
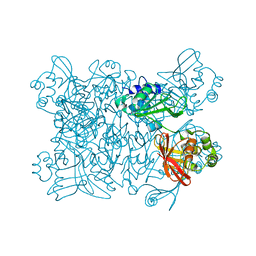 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3FE6
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand at pH 5.5 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
3FE9
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 7.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
1RP1
 
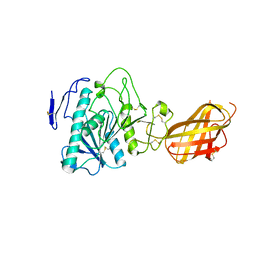 | | DOG PANCREATIC LIPASE RELATED PROTEIN 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PANCREATIC LIPASE RELATED PROTEIN 1 | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reactivation of the totally inactive pancreatic lipase RP1 by structure-predicted point mutations.
Proteins, 32, 1998
|
|
3FE8
 
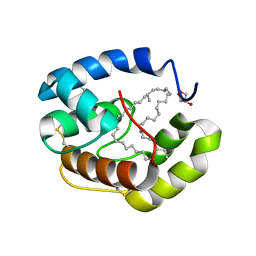 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 4.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
1CUS
 
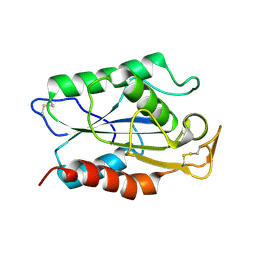 | |
1LPA
 
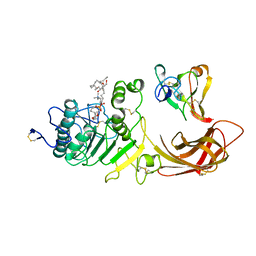 | | INTERFACIAL ACTIVATION OF THE LIPASE-PROCOLIPASE COMPLEX BY MIXED MICELLES REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, COLIPASE, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Van Tilbeurgh, H, Egloff, M.-P, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography.
Nature, 362, 1993
|
|
1LPB
 
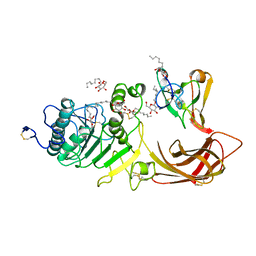 | | THE 2.46 ANGSTROMS RESOLUTION STRUCTURE OF THE PANCREATIC LIPASE COLIPASE COMPLEX INHIBITED BY A C11 ALKYL PHOSPHONATE | | Descriptor: | CALCIUM ION, COLIPASE, LIPASE, ... | | Authors: | Egloff, M.-P, Van Tilbeurgh, H, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate.
Biochemistry, 34, 1995
|
|
1G1K
 
 | | COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDING PROTEIN | | Authors: | Spinelli, S, Fierobe, H.-P, Belaich, A, Belaich, J.-P, Henrissat, B, Cambillau, C. | | Deposit date: | 2000-10-12 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition.
J.Mol.Biol., 304, 2000
|
|
1FWY
 
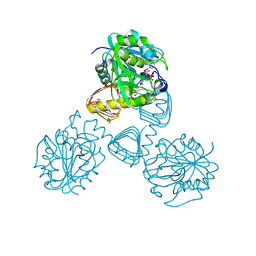 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE BOUND TO UDP-GLCNAC | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE, ... | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-25 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
2XF6
 
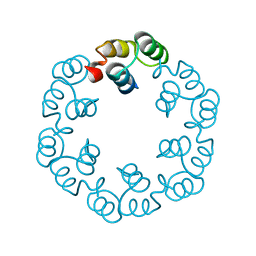 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
3D73
 
 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, at pH 7.0 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
1FXJ
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-26 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
3D78
 
 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D75
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, at pH 5.5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
