4J9B
 
 | |
4J9C
 
 | |
6F9Z
 
 | |
6F1M
 
 | |
6FA0
 
 | |
6F9Y
 
 | |
6F1L
 
 | |
6F1P
 
 | |
6F1O
 
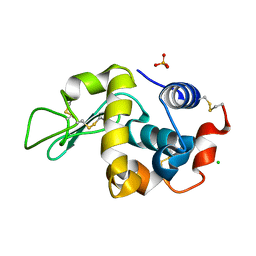 | | Orthorhombic Lysozyme crystallized at 298 K and pH 4.5 | | Descriptor: | CHLORIDE ION, Lysozyme C, PHOSPHATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Orthorhombic lysozyme crystallization at acidic pH values driven by phosphate binding.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6F9X
 
 | |
6F1R
 
 | |
4JZ3
 
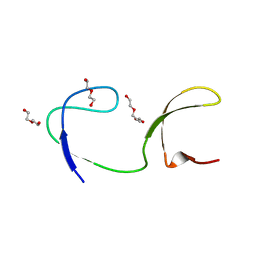 | | Crystal structure of the chicken c-Src-SH3 domain intertwined dimer | | Descriptor: | DI(HYDROXYETHYL)ETHER, Proto-oncogene tyrosine-protein kinase Src, TRIETHYLENE GLYCOL | | Authors: | Camara-Artigas, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
4JZ4
 
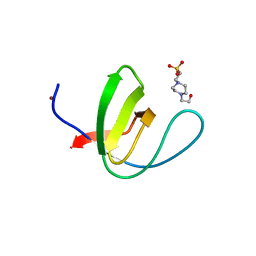 | | Crystal structure of chicken c-Src-SH3 domain: monomeric form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Camara-Artigas, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
4LE9
 
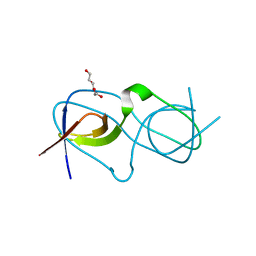 | | Crystal structure of a chimeric c-Src-SH3 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, TRIETHYLENE GLYCOL | | Authors: | Camara-Artigas, A, Martinez-Rodriguez, S, Ortiz-Salmeron, E, Martin-Garcia, J.M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | 3D domain swapping in a chimeric c-Src SH3 domain takes place through two hinge loops.
J.Struct.Biol., 186, 2014
|
|
7A2Y
 
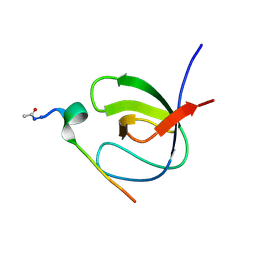 | |
7A2Z
 
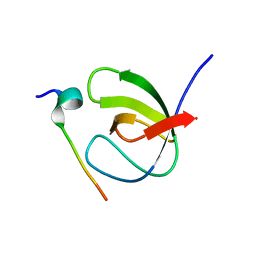 | |
7A2X
 
 | |
7A2S
 
 | |
7A2R
 
 | |
7A2V
 
 | |
7A2W
 
 | |
7A36
 
 | |
7A39
 
 | |
7A2T
 
 | |
7A31
 
 | |
