1Z7W
 
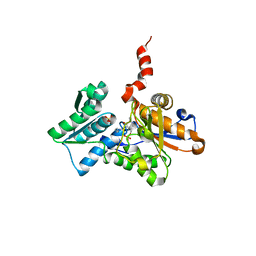 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Arabidopsis thaliana | | Descriptor: | Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Bonner, E.R, Cahoon, R.E, Knapke, S.M, Jez, J.M. | | Deposit date: | 2005-03-28 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of Cysteine Biosynthesis in Plants: STRUCTURAL AND FUNCTIONAL ANALYSIS OF O-ACETYLSERINE SULFHYDRYLASE FROM ARABIDOPSIS THALIANA.
J.Biol.Chem., 280, 2005
|
|
1Z7Y
 
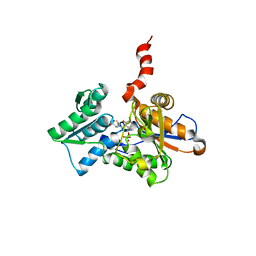 | | Crystal Structure of the Arabidopsis thaliana O-Acetylserine Sulfhydrylase K46A mutant | | Descriptor: | Cysteine synthase, N-[(3-HYDROXY-2-METHYL-5-{[(TRIHYDROXYPHOSPHORANYL)OXY]METHYL}PYRIDIN-4-YL)METHYLENE]METHIONINE | | Authors: | Bonner, E.R, Cahoon, R.E, Knapke, S.M, Jez, J.M. | | Deposit date: | 2005-03-28 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis of Cysteine Biosynthesis in Plants: STRUCTURAL AND FUNCTIONAL ANALYSIS OF O-ACETYLSERINE SULFHYDRYLASE FROM ARABIDOPSIS THALIANA.
J.Biol.Chem., 280, 2005
|
|
2VDJ
 
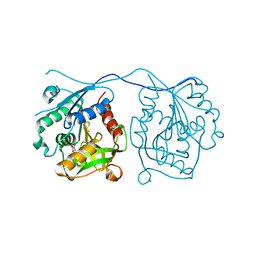 | | Crystal Structure of Homoserine O-acetyltransferase (metA) from Bacillus Cereus with Homoserine | | Descriptor: | HOMOSERINE O-SUCCINYLTRANSFERASE, L-HOMOSERINE, SULFATE ION | | Authors: | Zubieta, C, Arkus, K.A.J, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Single Amino Acid Change is Responsible for Evolution of Acyltransferase Specificity in Bacterial Methionine Biosynthesis.
J.Biol.Chem., 283, 2008
|
|
3KAJ
 
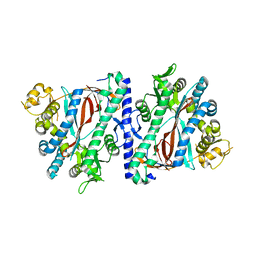 | | Apoenzyme structure of homoglutathione synthetase from Glycine max in open conformation | | Descriptor: | Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAL
 
 | | Structure of homoglutathione synthetase from Glycine max in closed conformation with homoglutathione, ADP, a sulfate ion, and three magnesium ions bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-gamma-glutamyl-L-cysteinyl-beta-alanine, MAGNESIUM ION, ... | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAK
 
 | | Structure of homoglutathione synthetase from Glycine max in open conformation with gamma-glutamyl-cysteine bound. | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
