2N62
 
 | | ddFLN5+110 | | Descriptor: | gelation factor, secretion monitor chimera | | Authors: | Cabrita, L.D, Cassaignau, A.M.E, Launay, H.M.M, Waudby, C.A, Camilloni, C, Robertson, A.L, Wang, X, Wlodarski, T, Wentink, A.S, Vendruscolo, M, Dobson, C.M, Christodoulou, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6G4A
 
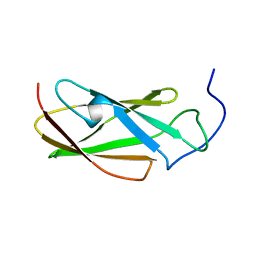 | | FLN5 (full length) | | Descriptor: | Gelation factor | | Authors: | Waudby, C.A, Wlodarski, T, Karyadi, M.-E, Cassaignau, A.M.E, Chan, S.H.S, Wentink, A.S, Schmidt-Engler, J.M, Camilloni, C, Vendruscolo, M, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mapping energy landscapes of a growing filamin domain reveals an intermediate associated with proline isomerization during biosynthesis
To Be Published
|
|
1SNG
 
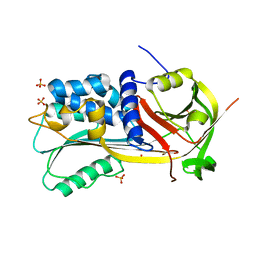 | | Structure of a Thermophilic Serpin in the Native State | | Descriptor: | COG4826: Serine protease inhibitor, SULFATE ION | | Authors: | Fulton, K.F, Buckle, A.M, Cabrita, L.D, Irving, J.A, Butcher, R.E, Smith, I, Reeve, S, Lesk, A.M, Bottomley, S.P, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-03-10 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition.
J.Biol.Chem., 280, 2005
|
|
1MTP
 
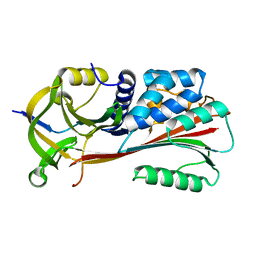 | | The X-ray crystal structure of a serpin from a thermophilic prokaryote | | Descriptor: | Serine Proteinase Inhibitor (SERPIN), Chain A, Chain B | | Authors: | Irving, J.A, Cabrita, L.D, Rossjohn, J, Pike, R.N, Bottomley, S.P, Whisstock, J.C. | | Deposit date: | 2002-09-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Structure, 11, 2003
|
|
7Z20
 
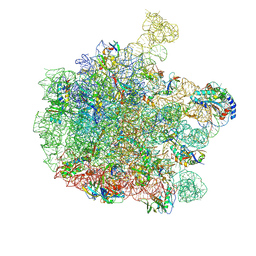 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZP8
 
 | | 70S E. coli ribosome with a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Chan, S.H.S, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZQ5
 
 | | 70S E. coli ribosome with truncated uL23 and uL24 loops | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZQ6
 
 | | 70S E. coli ribosome with truncated uL23 and uL24 loops and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZOD
 
 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
2MHQ
 
 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
2MHP
 
 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
