6DS8
 
 | |
6SPV
 
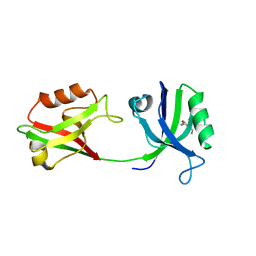 | | Crystal structure of PDZ1-2 from PSD-95 | | Descriptor: | Disks large homolog 4, GLUTATHIONE | | Authors: | Rodzli, N, Levy, C.W, Prince, S.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Dual PDZ Domain from Postsynaptic Density Protein 95 Forms a Scaffold with Peptide Ligand.
Biophys.J., 119, 2020
|
|
6XL0
 
 | | Caulobacter crescentus FljK filament | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
3D10
 
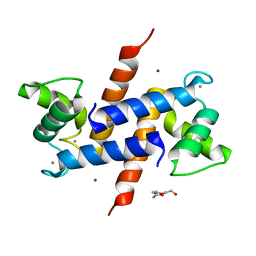 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 10.0 | | Descriptor: | CALCIUM ION, Protein S100-B, TRIETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
7W5R
 
 | | KRAS G12V and peptide complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, LEU-TYR-ASP-VAL-ALA, ... | | Authors: | Kim, H.J, Han, C.W, Jang, S.B. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structural basis of the oncogenic KRAS mutant and GJ101 complex.
Biochem.Biophys.Res.Commun., 641, 2023
|
|
7W5Q
 
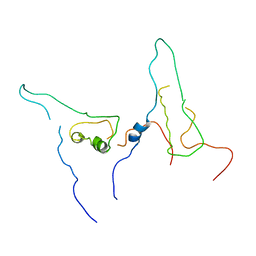 | |
6XKY
 
 | | Caulobacter crescentus FljK filament, straightened | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
7UBZ
 
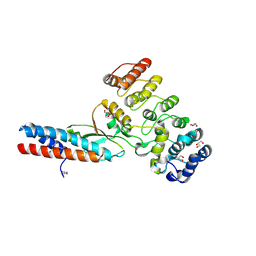 | | Chymotrypsin digested toxin/immunity complex for a T6SS lipase effector from E. cloacae | | Descriptor: | Ankyrin repeat domain-containing protein, CITRATE ANION, GLYCEROL, ... | | Authors: | Cuthbert, B.J, Jensen, S.J, Goulding, C.W, Hayes, C.S. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Toxin/immunity complex for a T6SS lipase effector from E. cloacae
To Be Published
|
|
6XD1
 
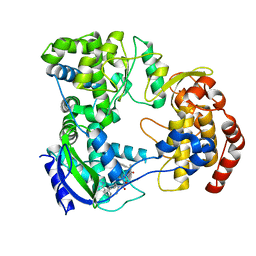 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-640 | | Descriptor: | (2R)-4-(butyl{[2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}carbamoyl)-1-(2,2-diphenylpropanoyl)piperazine-2-carboxylic acid, RNA-dependent RNA polymerase, ZINC ION | | Authors: | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
7UQ0
 
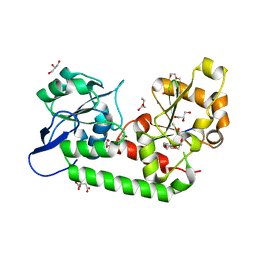 | | Putative periplasmic iron siderophore binding protein FecB (Rv3044) from Mycobacterium tuberculosis | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Chao, A, Cuthbert, B.J, Goulding, C.W. | | Deposit date: | 2022-04-18 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differentiating the roles of Mycobacterium tuberculosis substrate binding proteins, FecB and FecB2, in iron uptake.
Plos Pathog., 19, 2023
|
|
6DS7
 
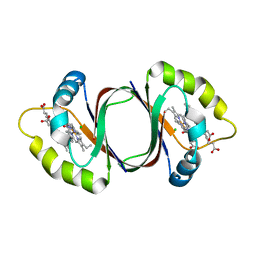 | |
7X7G
 
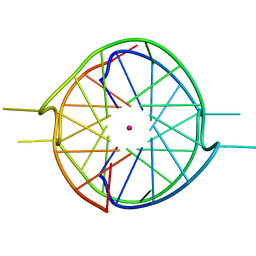 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7XDH
 
 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
6DZH
 
 | | HRAS G13D bound to GDP (H13GDP) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, GLYCEROL, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-04 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6EQG
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P21 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3J45
 
 | | Structure of a non-translocating SecY protein channel with the 70S ribosome | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L23, 50S ribosomal protein L24, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
3J46
 
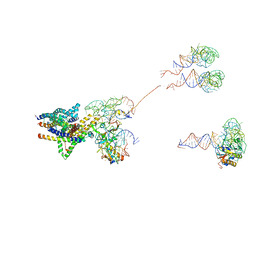 | | Structure of the SecY protein translocation channel in action | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L23P, ... | | Authors: | Akey, C.W, Park, E, Menetret, J.F, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-23 | | Last modified: | 2019-07-03 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
4HNY
 
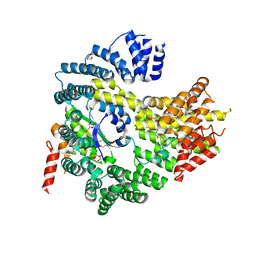 | | Apo N-terminal acetyltransferase complex A | | Descriptor: | GLYCEROL, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4EYY
 
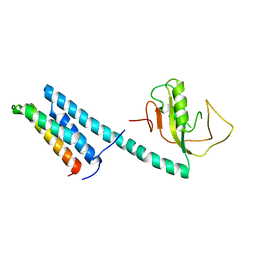 | | Crystal Structure of the IcmR-IcmQ complex from Legionella pneumophila | | Descriptor: | IcmQ, IcmR | | Authors: | Farelli, J.D, Gumbart, J, Akey, I.V, Amyot, W, Hempstead, A.D, Head, J.F, McKnight, C.J, Isberg, R.R, Akey, C.W. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | IcmQ in the Type 4b secretion system contains an NAD+ binding domain.
Structure, 21, 2013
|
|
4H8Q
 
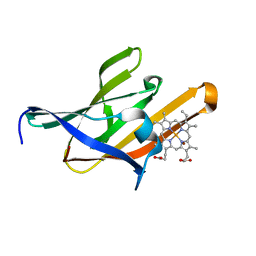 | | Structure of the Q29T IsdX2-NEAT5 mutant in complex with heme | | Descriptor: | Iron Transport-associated domain protein, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Owens, C.P, Goulding, C.W. | | Deposit date: | 2012-09-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Near-iron Transporter (NEAT) Domains of the Anthrax Hemophore IsdX2 Require a Critical Glutamine to Extract Heme from Methemoglobin.
J.Biol.Chem., 288, 2013
|
|
4H8P
 
 | | NEAT5 domain of IsdX2, a B. anthracis hemophore in complex with heme | | Descriptor: | Iron Transport-associated domain protein, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Owens, C.P, Goulding, C.W. | | Deposit date: | 2012-09-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Near-iron Transporter (NEAT) Domains of the Anthrax Hemophore IsdX2 Require a Critical Glutamine to Extract Heme from Methemoglobin.
J.Biol.Chem., 288, 2013
|
|
4HCT
 
 | | Crystal structure of ITK in complex with compound 52 | | Descriptor: | 3-{1-[(3R)-1-acryloylpiperidin-3-yl]-4-amino-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-(3-tert-butylphenyl)benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Zapf, C.W, Gerstenberger, B.S, Xing, L, Limburg, D.C, Anderson, D.R, Caspers, N, Han, S, Aulabaugh, A, Kurumbail, R, Shakya, S, Li, X, Spaulding, V, Czerwinski, R.M, Seth, N, Medley, Q.G. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
3FXE
 
 | |
4HNW
 
 | | The NatA Acetyltransferase Complex Bound To Inositol Hexakisphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
3FXD
 
 | |
