1BQO
 
 | | DISCOVERY OF POTENT, ACHIRAL MATRIX METALLOPROTEINASE INHIBITORS | | Descriptor: | 1,3-BIS-(4-METHOXY-BENZENESULFONYL)-5,5-DIMETHYL-HEXAHYDRO-PYRIMIDINE-2-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Pikul, S, Dunham, K.L.M, Almstead, N.G, De, B, Natchus, M.G, Anastasio, M.V, Mcphail, S.J, Snider, C.E, Taiwo, Y.O, Rydel, T.J, Dunaway, C.M, Gu, F, Mieling, G.E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent, achiral matrix metalloproteinase inhibitors.
J.Med.Chem., 41, 1998
|
|
1EFA
 
 | |
1C1K
 
 | | BACTERIOPHAGE T4 GENE 59 HELICASE ASSEMBLY PROTEIN | | Descriptor: | BPT4 GENE 59 HELICASE ASSEMBLY PROTEIN, CHLORIDE ION, IRIDIUM ION | | Authors: | Mueser, T.C, Jones, C.E, Nossal, N.G, Hyde, C.C. | | Deposit date: | 1999-07-22 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bacteriophage T4 gene 59 helicase assembly protein binds replication fork DNA. The 1.45 A resolution crystal structure reveals a novel alpha-helical two-domain fold.
J.Mol.Biol., 296, 2000
|
|
1BU6
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
1D1J
 
 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
1F39
 
 | | CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR C-TERMINAL DOMAIN | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Bell, C.E, Frescura, P, Hochschild, A, Lewis, M. | | Deposit date: | 2000-06-01 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding.
Cell(Cambridge,Mass.), 101, 2000
|
|
1F7S
 
 | | CRYSTAL STRUCTURE OF ADF1 FROM ARABIDOPSIS THALIANA | | Descriptor: | ACTIN DEPOLYMERIZING FACTOR (ADF), LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Bowman, G.D, Nodelman, I.M, Lindberg, U, Chua, N.H, Schutt, C.E. | | Deposit date: | 2000-06-27 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comparative structural analysis of the ADF/cofilin family.
Proteins, 41, 2000
|
|
1E5G
 
 | | Solution structure of central CP module pair of a pox virus complement inhibitor | | Descriptor: | COMPLEMENT CONTROL PROTEIN C3 | | Authors: | Henderson, C.E, Bromek, K, Mullin, N.P, Smith, B.O, Uhrin, D, Barlow, P.N. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-31 | | Last modified: | 2013-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Central Ccp Module Pair of a Poxvirus Complement Control Protein
J.Mol.Biol., 307, 2001
|
|
1FHM
 
 | | X-RAY CRYSTAL STRUCTURE OF REDUCED RUBREDOXIN | | Descriptor: | FE (II) ION, RUBREDOXIN | | Authors: | Min, T, Ergenekan, C.E, Eidsness, M.K, Ichiye, T, Kang, C. | | Deposit date: | 2000-08-02 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin.
Protein Sci., 10, 2001
|
|
1FHH
 
 | | X-RAY CRYSTAL STRUCTURE OF OXIDIZED RUBREDOXIN | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Ergenekan, C.E, Eidsness, M.K, Ichiye, T, Kang, C. | | Deposit date: | 2000-08-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin.
Protein Sci., 10, 2001
|
|
1PGP
 
 | | CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, 6-PHOSPHOGLUCONIC ACID, SULFATE ION | | Authors: | Adams, M.J, Phillips, C, Gover, S, Naylor, C.E. | | Deposit date: | 1994-07-18 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
Structure, 2, 1994
|
|
1SFJ
 
 | | 2.4A Crystal structure of Staphylococcus aureus type I 3-dehydroquinase, with 3-dehydroquinate bound | | Descriptor: | 3-DEHYDROSHIKIMATE, 3-dehydroquinate dehydratase | | Authors: | Nichols, C.E, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Staphylococcus aureus type I dehydroquinase from enzyme turnover experiments
Proteins, 56, 2004
|
|
8C8J
 
 | | Long Interspersed Nuclear Element 1 (LINE-1) reverse transcriptase ternary complex with hybrid duplex and dTTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Walpole, T.B, Baldwin, E. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
1SR4
 
 | | Crystal Structure of the Haemophilus ducreyi cytolethal distending toxin | | Descriptor: | BROMIDE ION, Cytolethal distending toxin subunit A, cytolethal distending toxin protein B, ... | | Authors: | Nesic, D, Hsu, Y, Stebbins, C.E. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assembly and Function of a Bacterial Genotoxin
Nature, 429, 2004
|
|
1SFL
 
 | | 1.9A Crystal structure of Staphylococcus aureus type I 3-dehydroquinase, apo form | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Nichols, C.E, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Staphylococcus aureus type I dehydroquinase from enzyme turnover experiments.
Proteins, 56, 2004
|
|
1Q8L
 
 | | Second Metal Binding Domain of the Menkes ATPase | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Jones, C.E, Daly, N.L, Cobine, P.A, Craik, D.J, Dameron, C.T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and metal binding studies of the second copper binding domain of the Menkes ATPase.
J.Struct.Biol., 143, 2003
|
|
1Q08
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator, at 1.9 A resolution (space group P212121) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q05
 
 | | Crystal structure of the Cu(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | COPPER (I) ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q0A
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group C222) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q07
 
 | | Crystal structure of the Au(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | GOLD ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1S73
 
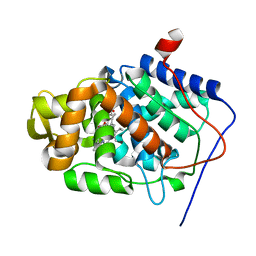 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (R-isomer) [MpCcP-R] | | Descriptor: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER | | Authors: | Bhaskar, B, Immoos, C.E, Sulc, F, Choen, M.S, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2004-01-28 | | Release date: | 2005-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structures of reduced, resting and NO-bound states of mesopone cytochorme c peroxidase (MpCcP) (R-isomer)
To be Published
|
|
1SBM
 
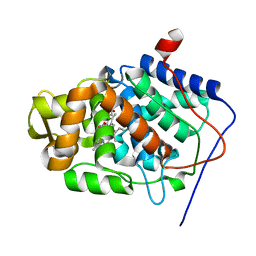 | | Crystal Structure of Reduced Mesopone cytochrome c peroxidase (R-isomer) | | Descriptor: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER | | Authors: | Bhaskar, B, Immoos, C.E, Sulc, F, Cohen, M.S, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2004-02-10 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structures of Resting (Fe3+), Reduced (Fe2+) and Reduced-NO adduct of Mesopone cytochrome c peroxidase (MpCcP) - R-isomer
To be Published
|
|
1Q09
 
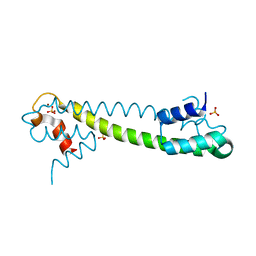 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q5Z
 
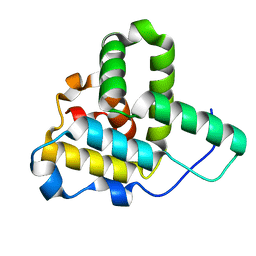 | | Crystal Structure of the C-terminal Actin Binding Domain of Salmonella Invasion Protein A (SipA) | | Descriptor: | SipA | | Authors: | Stebbins, C.E, Lilic, M, Galkin, V.E, Orlova, A, VanLoock, M.S, Egelman, E.H. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms.
Science, 301, 2003
|
|
1Q06
 
 | | Crystal structure of the Ag(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | SILVER ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
