2H7V
 
 | | Co-crystal structure of YpkA-Rac1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Migration-inducing protein 5, ... | | Authors: | Prehna, G, Ivanov, M, Bliska, J.B, Stebbins, C.E. | | Deposit date: | 2006-06-04 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Yersinia virulence depends on mimicry of host rho-family nucleotide dissociation inhibitors.
Cell(Cambridge,Mass.), 126, 2006
|
|
1Q09
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q5Z
 
 | | Crystal Structure of the C-terminal Actin Binding Domain of Salmonella Invasion Protein A (SipA) | | Descriptor: | SipA | | Authors: | Stebbins, C.E, Lilic, M, Galkin, V.E, Orlova, A, VanLoock, M.S, Egelman, E.H. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms.
Science, 301, 2003
|
|
2GGN
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
2FYQ
 
 | |
2GEM
 
 | |
2GHK
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
1SDQ
 
 | | Structure of reduced-NO adduct of mesopone cytochrome c peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER, ... | | Authors: | Bhaskar, B, Immoos, C.E, Sulc, F, Cohem, M.S, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2004-02-13 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of resting (Fe3+), reduced (Fe2+) and NO-bound states of mesopone cytochrome c peroxidase (MpCcP) (R-isomer)
To be Published
|
|
1SG6
 
 | | Crystal structure of Aspergillus nidulans 3-dehydroquinate synthase (AnDHQS) in complex with Zn2+ and NAD+, at 1.7D | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentafunctional AROM polypeptide, ZINC ION | | Authors: | Nichols, C.E, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the 'open' form of Aspergillus nidulans 3-dehydroquinate synthase at 1.7 A resolution from crystals grown following enzyme turnover.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2HFN
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | FLAVIN MONONUCLEOTIDE, Synechocystis Photoreceptor (Slr1694) | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
1SGK
 
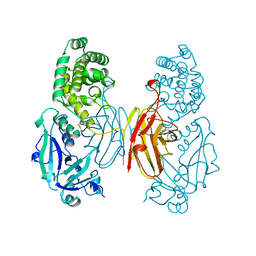 | | NUCLEOTIDE-FREE DIPHTHERIA TOXIN | | Descriptor: | DIPHTHERIA TOXIN (DIMERIC) | | Authors: | Bell, C.E, Eisenberg, D. | | Deposit date: | 1996-09-12 | | Release date: | 1996-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of nucleotide-free diphtheria toxin.
Biochemistry, 36, 1997
|
|
1PGP
 
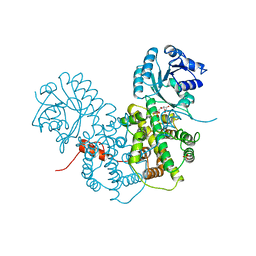 | | CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, 6-PHOSPHOGLUCONIC ACID, SULFATE ION | | Authors: | Adams, M.J, Phillips, C, Gover, S, Naylor, C.E. | | Deposit date: | 1994-07-18 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
Structure, 2, 1994
|
|
2HNY
 
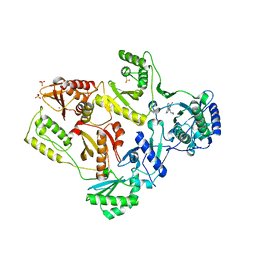 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HFO
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | Activator of photopigment and puc expression, FLAVIN MONONUCLEOTIDE | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
2HND
 
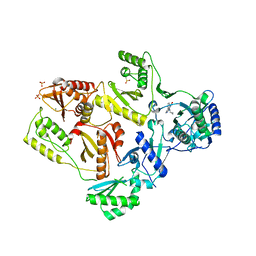 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HO0
 
 | |
2FM8
 
 | |
2HKC
 
 | | NMR Structure of the IQ-modified Dodecamer CTCGGC[IQ]GCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
2HNF
 
 | |
2HKB
 
 | | NMR Structure of the B-DNA Dodecamer CTCGGCGCCATC | | Descriptor: | 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
1Q8L
 
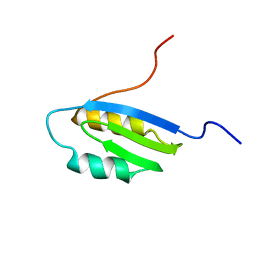 | | Second Metal Binding Domain of the Menkes ATPase | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Jones, C.E, Daly, N.L, Cobine, P.A, Craik, D.J, Dameron, C.T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and metal binding studies of the second copper binding domain of the Menkes ATPase.
J.Struct.Biol., 143, 2003
|
|
2DPG
 
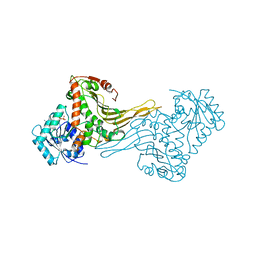 | | COMPLEX OF INACTIVE MUTANT (H240->N) OF GLUCOSE 6-PHOSPHATE DEHYDROGENASE FROM LEUCONOSTOC MESENTEROIDES WITH NADP+ | | Descriptor: | GLUCOSE 6-PHOSPHATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Adams, M.J, Naylor, C.E, Paludin, S, Gover, S. | | Deposit date: | 1998-04-17 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | On the mechanism of the reaction catalyzed by glucose 6-phosphate dehydrogenase.
Biochemistry, 37, 1998
|
|
1QCW
 
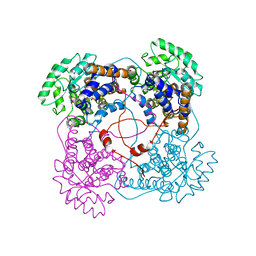 | | Flavocytochrome B2, ARG289LYS mutant | | Descriptor: | FLAVOCYTOCHROME B2, N-SULFO-FLAVIN MONONUCLEOTIDE | | Authors: | Mowat, C.G, Durley, R.C.E, Pike, A.D, Barton, J.D, Chen, Z.-W, Mathews, F.S, Lederer, F, Reid, G.A, Chapman, S.K. | | Deposit date: | 1999-05-07 | | Release date: | 1999-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Kinetic and crystallographic studies on the active site Arg289Lys mutant of flavocytochrome b2 (yeast L-lactate dehydrogenase)
Biochemistry, 39, 2000
|
|
2EEW
 
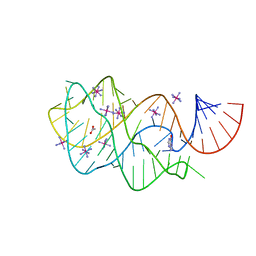 | | Guanine Riboswitch U47C mutant bound to hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), Guanine riboswitch, ... | | Authors: | Gilbert, S.D, Love, C.E, Batey, R.T. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutational analysis of the purine riboswitch aptamer domain
Biochemistry, 46, 2007
|
|
1T3D
 
 | | Crystal structure of Serine Acetyltransferase from E.coli at 2.2A | | Descriptor: | CYSTEINE, Serine acetyltransferase | | Authors: | Pye, V.E, Tingey, A.P, Robson, R.L, Moody, P.C.E. | | Deposit date: | 2004-04-26 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure and Mechanism of Serine Acetyltransferase from Escherichia coli
J.Biol.Chem., 279, 2004
|
|
