6G0B
 
 | |
6G09
 
 | | Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6DH4
 
 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6G4A
 
 | | FLN5 (full length) | | Descriptor: | Gelation factor | | Authors: | Waudby, C.A, Wlodarski, T, Karyadi, M.-E, Cassaignau, A.M.E, Chan, S.H.S, Wentink, A.S, Schmidt-Engler, J.M, Camilloni, C, Vendruscolo, M, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mapping energy landscapes of a growing filamin domain reveals an intermediate associated with proline isomerization during biosynthesis
To Be Published
|
|
6FYZ
 
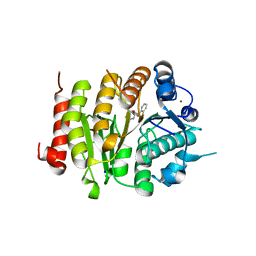 | | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor | | Descriptor: | (2~{S})-2-(2-fluorophenyl)-2-[4-(2-methylpyrimidin-5-yl)phenyl]-~{N}-oxidanyl-ethanamide, Histone deacetylase 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Maillard, M.C, Dominguez, C. | | Deposit date: | 2018-03-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6E3Y
 
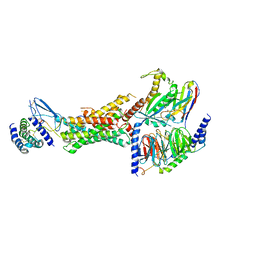 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
6E47
 
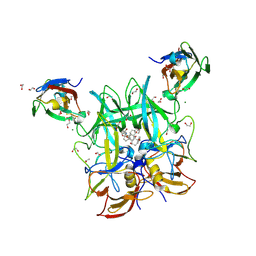 | |
6E48
 
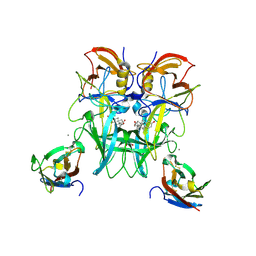 | | Crystal Structure of the Murine Norovirus VP1 P domain in complex with the CD300lf Receptor and Lithocholic Acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G0N
 
 | | Crystal Structure of a GH8 catalytic mutant xylohexaose complex xylanase from Teredinibacter turnerae | | Descriptor: | GLYCEROL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose, ... | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G81
 
 | | Solution structure of the Ni metallochaperone HypA from Helicobacter pylori | | Descriptor: | Hydrogenase maturation factor HypA, ZINC ION | | Authors: | Spronk, C.A.E.M, Zerko, S, Gorka, M, Kozminski, W, Bardiaux, B, Zambelli, B, Musiani, F, Piccioli, M, Hu, H, Maroney, M, Ciurli, S. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools.
J. Biol. Inorg. Chem., 23, 2018
|
|
6GIZ
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GO3
 
 | | TdT chimera (Loop1 of pol mu) - apoenzyme | | Descriptor: | DNA nucleotidylexotransferase,DNA-directed DNA/RNA polymerase mu,DNA nucleotidylexotransferase | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6EC3
 
 | | Crystal Structure of EvdMO1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Methyltransferase domain-containing protein, NICKEL (II) ION | | Authors: | McCulloch, K.M, Iverson, T.M, Starbird, C.A, Perry, N.A, Chen, Q, Berndt, S, Yamakawa, I, Loukachevitch, L.V. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
6G00
 
 | | Crystal Structure of a GH8 xylanase from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, SODIUM ION | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6GJ9
 
 | |
6GO4
 
 | | TdT chimera (Loop1 of pol mu) - binary complex with ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA nucleotidylexotransferase,DNA-directed DNA/RNA polymerase mu,DNA nucleotidylexotransferase, MAGNESIUM ION, ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6GIT
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - PRODUCT COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GFG
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with D-chiro-IP6 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-chiro inositol hexakisphosphate, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6GO7
 
 | | TdT chimera (Loop1 of pol mu) - full DNA synapsis complex | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*AP*AP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*C)-3'), ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6GYN
 
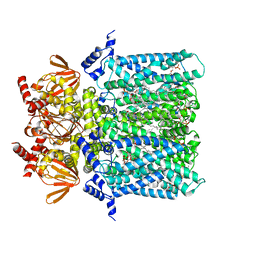 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
6GYO
 
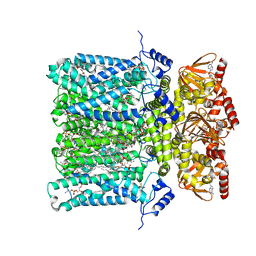 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel in complex with cAMP | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
6GJA
 
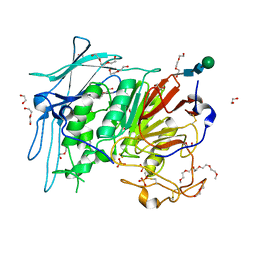 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - H229A MUTANT | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GO5
 
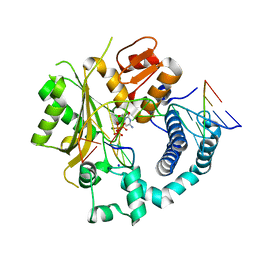 | | TdT chimera (Loop1 of pol mu) - Ternary complex with 1-nt gapped DNA substrate | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(*AP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*GP*GP*CP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6GFH
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with neo-IP5 and ATP | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
