6NC2
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody ACS202 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Yuan, M, Copps, J, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
6NIV
 
 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
6NK6
 
 | | Electron Cryo-Microscopy Of Chikungunya VLP in complex with mouse Mxra8 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Basore, K, Kim, A.S, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor.
Cell, 177, 2019
|
|
2AWI
 
 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.k, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1CDE
 
 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | 5-DEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | Deposit date: | 1992-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1RTP
 
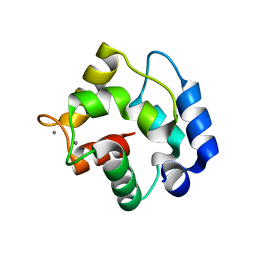 | | REFINED X-RAY STRUCTURE OF RAT PARVALBUMIN, A MAMMALIAN ALPHA-LINEAGE PARVALBUMIN, AT 2.0 A RESOLUTION | | Descriptor: | ALPHA-PARVALBUMIN, CALCIUM ION | | Authors: | Mcphalen, C.A, Sielecki, A.R, Santarsiero, B.D, James, M.N.G. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of rat parvalbumin, a mammalian alpha-lineage parvalbumin, at 2.0 A resolution.
J.Mol.Biol., 235, 1994
|
|
2VJF
 
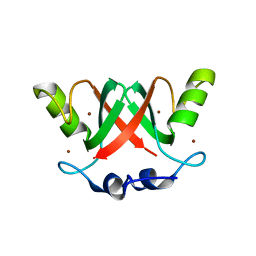 | | Crystal Structure of the MDM2-MDMX RING Domain Heterodimer | | Descriptor: | CITRATE ANION, E3 UBIQUITIN-PROTEIN LIGASE MDM2, MDM4 PROTEIN, ... | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mdm2/Mdmx Ring Domain Heterodimer Reveals Dimerization is Required for Their Ubiquitylation in Trans.
Cell Death Differ., 15, 2008
|
|
6N6X
 
 | |
2AGJ
 
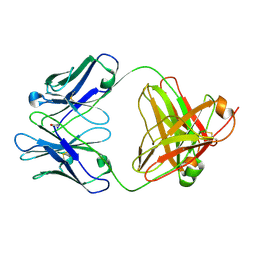 | | Crystal Structure of a glycosylated Fab from an IgM cryoglobulin with properties of a natural proteolytic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Yvo Fab, Heavy Chain, ... | | Authors: | Ramsland, P.A, Terzyan, S.S, Cloud, G, Bourne, C.R, Farrugia, W, Tribbick, G, Geysen, H.M, Moomaw, C.R, Slaughter, C.A, Edmundson, A.B. | | Deposit date: | 2005-07-27 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a glycosylated Fab from an IgM cryoglobulin with properties of a natural proteolytic antibody
Biochem.J., 395, 2006
|
|
6N09
 
 | |
2WLY
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chitinase inhibition by chitobiose and chitotriose thiazolines.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
1TKL
 
 | | Yeast Oxygen-Dependent Coproporphyrinogen Oxidase | | Descriptor: | Coproporphyrinogen III oxidase | | Authors: | Phillips, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxygen-dependant Coproporphyrinogen Oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
2AXU
 
 | | Structure of PrgX | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1SYS
 
 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | Descriptor: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
6MX0
 
 | | Crystal structure of human STING apoprotein (G230A, H232R, R293Q) | | Descriptor: | CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A, Siu, T, Ho, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING.
ACS Med Chem Lett, 10, 2019
|
|
2AZQ
 
 | | Crystal Structure of Catechol 1,2-Dioxygenase from Pseudomonas arvilla C-1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, FE (III) ION, catechol 1,2-dioxygenase | | Authors: | Earhart, C.A, Vetting, M.W, Gosu, R, Michaud-Soret, I, Que, L, Ohlendorf, D.H. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of catechol 1,2-dioxygenase from Pseudomonas arvilla
Biochem.Biophys.Res.Commun., 338, 2005
|
|
6N6W
 
 | | OXA-23 mutant F110A/M221A neutral pH form | | Descriptor: | Beta-lactamase oxa23 | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Role of the Hydrophobic Bridge in the Carbapenemase Activity of Class D beta-Lactamases.
Antimicrob. Agents Chemother., 63, 2019
|
|
2W3I
 
 | | Crystal Structure of FXa in complex with 4,4-disubstituted pyrrolidine-1,2-dicarboxamide inhibitor 2 | | Descriptor: | (2R,4S)-N^1^-(4-chlorophenyl)-4-(2,4-difluorophenyl)-4-hydroxy-N^2^-(2-oxo-2H-1,3'-bipyridin-6'-yl)pyrrolidine-1,2-dicarboxamide, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Zhang, E, Mochalkin, I, Casimiro-Garcia, A, Van Huis, C.A. | | Deposit date: | 2008-11-12 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploration of 4,4-Disubstituted Pyrrolidine-1,2-Dicarboxamides as Potent, Orally Active Factor Xa Inhibitors with Extended Duration of Action.
Bioorg.Med.Chem., 17, 2009
|
|
1CDD
 
 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | PHOSPHATE ION, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | Deposit date: | 1992-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2W2L
 
 | | Crystal structure of the holo forms of Rhodotorula graminis D- mandelate dehydrogenase at 2.5A. | | Descriptor: | D-MANDELATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Vachieri, S.G, Cole, A.R, Bagneris, C, Baker, D.P, Fewson, C.A, Basak, A.K. | | Deposit date: | 2008-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Apo and Holo Forms of Rhodotorula Graminis D(-)-Mandelate Dehydrogenase
To be Published
|
|
2VOS
 
 | | Mycobacterium tuberculosis Folylpolyglutamate synthase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, FOLYLPOLYGLUTAMATE SYNTHASE PROTEIN FOLC, ... | | Authors: | Young, P.G, Baker, E.N, Metcalf, P, Smith, C.A. | | Deposit date: | 2008-02-19 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Mycobacterium Tuberculosisfolylpolyglutamate Synthase Complexed with Adp and Amppcp.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6NIJ
 
 | | PGT145 Fab in complex with full length AMC011 HIV-1 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 Glycoprotein 120, ... | | Authors: | Cottrell, C.A, Torrents de la Pena, A, Rantalainen, K, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-29 | | Release date: | 2019-07-31 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
1TS2
 
 | | T128A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
5ND3
 
 | | Microtubule-bound MKLP2 motor domain in the with no nucleotide | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF20A, ... | | Authors: | Atherton, J, Yu, I.M, Cook, A, Muretta, J.M, Joseph, A.P, Major, J, Sourigues, Y, Clause, J, Topf, M, Rosenfeld, S.S, Houdusse, A, Moores, C.A. | | Deposit date: | 2017-03-07 | | Release date: | 2017-10-04 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry.
Elife, 6, 2017
|
|
2WK2
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline dithioamide. | | Descriptor: | 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose-(1-4)-2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHITINASE A, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
