4XA1
 
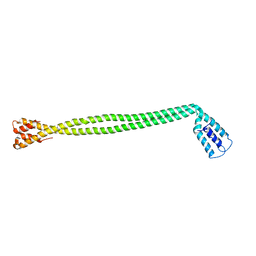 | | Crystal Structure of the coiled-coil surrounding Skip 1 of MYH7 | | Descriptor: | Gp7-MYH7(1173-1238)-EB1 chimera protein | | Authors: | Taylor, K.C, Buvoli, M, Korkmaz, E.N, Buvoli, A, Zheng, Y, Heinz, N.T, Qiang, C, Leinwand, L.A, Rayment, I. | | Deposit date: | 2014-12-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XA4
 
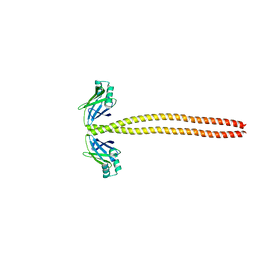 | | Crystal Structure of the coiled-coil surrounding Skip 3 of MYH7 | | Descriptor: | Xrcc4-MYH7(1551-1609) chimera protein | | Authors: | Taylor, K.C, Buvoli, M, Korkmaz, E.N, Buvoli, A, Zheng, Y, Heinz, N.T, Qiang, C, Leinwand, L.A, Rayment, I. | | Deposit date: | 2014-12-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XA3
 
 | | Crystal structure of the coiled-coil surrounding Skip 2 of MYH7 | | Descriptor: | Gp7-MYH7(1361-1425)-Eb1 chimera protein | | Authors: | Taylor, K.C, Buvoli, M, Korkmaz, E.N, Buvoli, A, Zheng, Y, Heinz, N.T, Qiang, C, Leinwand, L.A, Rayment, I. | | Deposit date: | 2014-12-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XA6
 
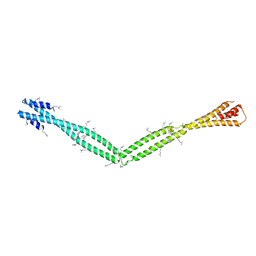 | | Crystal Structure of the coiled-coil surrounding Skip 4 of MYH7 | | Descriptor: | Gp7-MYH7(1777-1855)-EB1 chimera protein | | Authors: | Taylor, K.C, Buvoli, M, Korkmaz, E.N, Buvoli, A, Zheng, Y, Heinz, N.T, Qiang, C, Leinwand, L.A, Rayment, I. | | Deposit date: | 2014-12-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
