2KVQ
 
 | |
2LCL
 
 | |
4C00
 
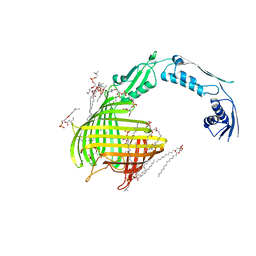 | | Crystal structure of TamA from E. coli | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Gruss, F, Zaehringer, F, Jakob, R.P, Burmann, B.M, Hiller, S, Maier, T. | | Deposit date: | 2013-07-30 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structural Basis of Autotransporter Translocation by Tama
Nat.Struct.Mol.Biol., 20, 2013
|
|
6YHZ
 
 | |
6YI2
 
 | |
2LQ8
 
 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
5OWJ
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 2) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
5OWI
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 1) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
4BZA
 
 | | Crystal structure of TamA POTRA domains 1-3 from E. coli | | Descriptor: | 1,2-ETHANEDIOL, TRANSLOCATION AND ASSEMBLY MODULE TAMA | | Authors: | Jakob, R.P, Gruss, F, Zaehringer, F, Burmann, B.M, Hiller, S, Maier, T. | | Deposit date: | 2013-07-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | The Structural Basis of Autotransporter Translocation by Tama
Nat.Struct.Mol.Biol., 20, 2013
|
|
