1LAP
 
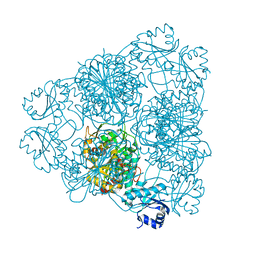 | | MOLECULAR STRUCTURE OF LEUCINE AMINOPEPTIDASE AT 2.7-ANGSTROMS RESOLUTION | | Descriptor: | Cytosol aminopeptidase, ZINC ION | | Authors: | Burley, S.K, David, P.R, Taylor, A, Lipscomb, W.N. | | Deposit date: | 1990-08-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular structure of leucine aminopeptidase at 2.7-A resolution.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1M6S
 
 | | Crystal Structure Of Threonine Aldolase | | Descriptor: | CALCIUM ION, CHLORIDE ION, L-allo-threonine aldolase | | Authors: | Burley, S.K, Kielkopf, C.L. | | Deposit date: | 2002-07-17 | | Release date: | 2002-12-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of Threonine Aldolase Complexes: Structural Basis of Substrate Recognition
Biochemistry, 41, 2002
|
|
1VH5
 
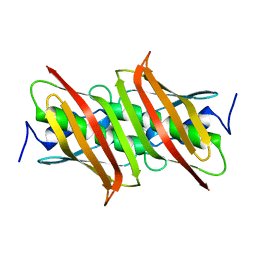 | |
1YR0
 
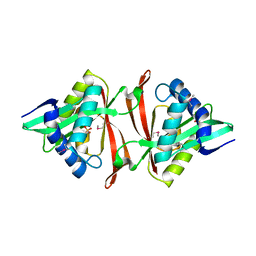 | |
1YW1
 
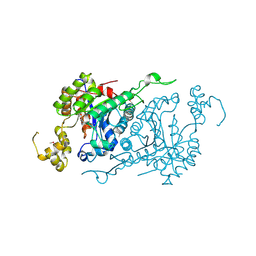 | |
4MZY
 
 | | Crystal structure of enterococcus faecalis nicotinate phosphoribosyltransferase with malonate and phosphate bound | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Wasserman, S.R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Enterococcus Faecalis Nicotinate Phosphoribosyltransferase
To be Published
|
|
4N7T
 
 | | Crystal structure of phosphorylated phosphopentomutase from streptococcus mutans | | Descriptor: | AZIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Bonanno, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structure of phosphorylated phosphopentomutase from streptococcus mutans
To be Published
|
|
1HU3
 
 | | MIDDLE DOMAIN OF HUMAN EIF4GII | | Descriptor: | EIF4GII | | Authors: | Marcotrigiano, J, Lomakin, I, Pestova, T.V, Hellen, C.U.T, Sonenberg, N, Burley, S.K. | | Deposit date: | 2001-01-03 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery.
Mol.Cell, 7, 2001
|
|
4RKQ
 
 | | Crystal structure of LacI family transcriptional regulator from Arthrobacter sp. FB24, target EFI-560007 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator PurR from Arthrobacter Sp, Target Nysgxrc 11027R
To be Published
|
|
4RKR
 
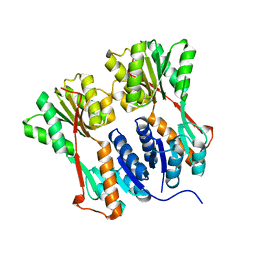 | | Crystal structure of LacI family transcriptional regulator from Arthrobacter sp. FB24, target EFI-560007, complex with lactose | | Descriptor: | Transcriptional regulator, LacI family, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator PurR from Arthrobacter Sp, Target Nysgxrc 11027R
To be Published
|
|
4D8L
 
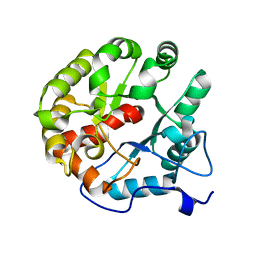 | | Crystal structure of the 2-pyrone-4,6-dicarboxylic acid hydrolase from sphingomonas paucimobilis | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DWD
 
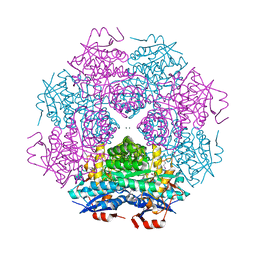 | | Crystal structure of mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans PD1222 complexed with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans PD1222 complexed with magnesium
To be Published
|
|
5Q0K
 
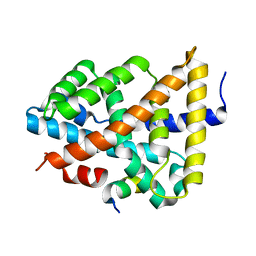 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0V
 
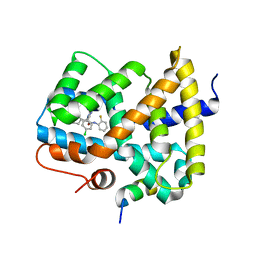 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-(2-fluorophenyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1A
 
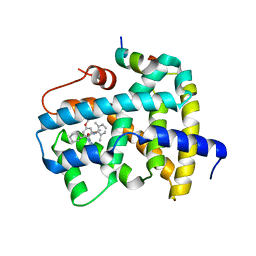 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,4-dimethoxyphenyl)-1H-benzimidazol-1-yl]-N-(2,6-dimethylphenyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0P
 
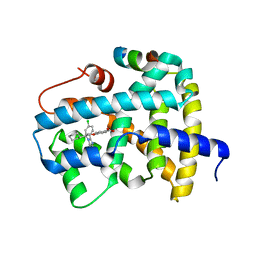 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q16
 
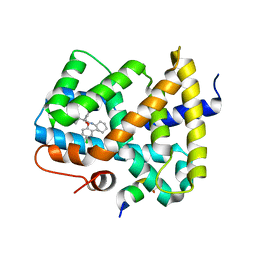 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[(4-chloro-2-methylphenoxy)methyl]-6-fluoro-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q19
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-[2-(2,4-dimethoxyphenyl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0R
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,1-dibenzyl-6-[(2-fluorophenyl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Z
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl (5S)-3-(3,4-difluorobenzene-1-carbonyl)-1,1-dimethyl-1,2,3,4,5,6-hexahydroazepino[4,5-b]indole-5-carboxylate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 3-(2-chlorophenyl)-N-[(1R)-1-(naphthalen-2-yl)ethyl]-5-(propan-2-yl)-1,2-oxazole-4-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5QBW
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-{[2-chloro-5-(2-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-3-oxo-2,3-dihydropyridazin-4-yl)phenyl]methyl}-4-fluorobenzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCB
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, tert-butyl 4-(2-{3-[3-{[(3-methylbut-2-enoyl)amino]methyl}-4-(trifluoromethyl)phenyl]-1-[3-(morpholin-4-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-oxoethyl)piperidine-1-carboxylate | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | Descriptor: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDC
 
 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | Descriptor: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
