5M2S
 
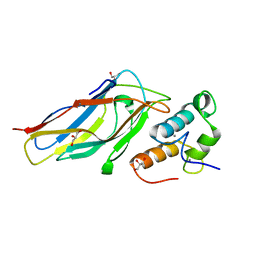 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Doc8: Type I dockerin repeat domain from family 9 glycoside hydrolase WP_009982745[Ruminococcus flavefaciens], GLYCEROL, ... | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
5AOZ
 
 | | High resolution SeMet structure of the third cohesin from Ruminococcus flavefaciens scaffoldin protein, ScaB | | Descriptor: | GLYCEROL, PUTATIVE CELLULOSOMAL SCAFFOLDIN PROTEIN | | Authors: | Bule, P, Carvalho, A.L, Santos, H, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural Characterization of the Third Cohesin from Ruminococcus Flavefaciens Scaffoldin Protein, Scab
To be Published
|
|
6S04
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-glycolylneuraminic acid | | Descriptor: | ACETATE ION, GLYCEROL, N-glycolyl-beta-neuraminic acid, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6S0F
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
5N5P
 
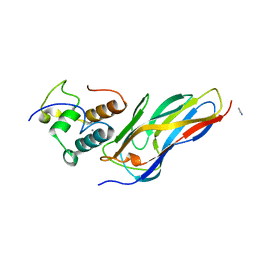 | | Crystal structure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A | | Descriptor: | ACETONITRILE, CALCIUM ION, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Carvalho, A.L, Najmudin, S, Fontes, C.M.G.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Higher order scaffoldin assembly in Ruminococcus flavefaciens cellulosome is coordinated by a discrete cohesin-dockerin interaction.
Sci Rep, 8, 2018
|
|
5NRK
 
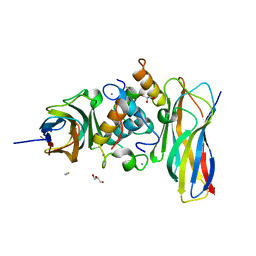 | | Crystal structure of the sixth cohesin from Acetivibrio cellulolyticus' scaffoldin B in complex with Cel5 dockerin S15I, I16N mutant | | Descriptor: | CALCIUM ION, DocCel5: Type I dockerin repeat domain from A. cellulolyticus family 5 endoglucanase WP_010249057 S15I, I16N mutant, ... | | Authors: | Bule, P, Najmudin, S, Fontes, C.M.G.A, Alves, V.D. | | Deposit date: | 2017-04-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
J. Biol. Chem., 293, 2018
|
|
5NRM
 
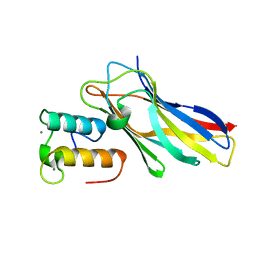 | | Crystal structure of the sixth cohesin from Acetivibrio cellulolyticus' scaffoldin B in complex with Cel5 dockerin S51I, L52N mutant | | Descriptor: | CALCIUM ION, DocCel5: Type I dockerin repeat domain from A. cellulolyticus family 5 endoglucanase WP_010249057 S51I, L52N mutant, ... | | Authors: | Bule, P, Najmudin, S, Fontes, C.M.G.A, Alves, V.D. | | Deposit date: | 2017-04-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
J. Biol. Chem., 293, 2018
|
|
6S00
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid | | Descriptor: | GLYCEROL, N-acetyl-beta-neuraminic acid, TETRAETHYLENE GLYCOL, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6S0E
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ACETATE ION, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
5M2O
 
 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Group I Dockerin, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
6RZD
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
7QUZ
 
 | | Crystal structure of the SeMet octameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, GLYCEROL | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-01-19 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Bacillus circulans IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
4UYP
 
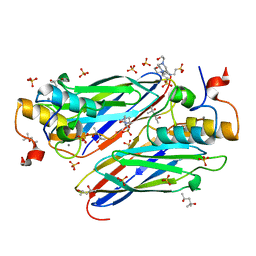 | | High resolution structure of the third cohesin ScaC in complex with the ScaB dockerin with a mutation in the N-terminal helix (IN to SI) from Acetivibrio cellulolyticus displaying a type I interaction. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Cameron, K, Alves, V.D, Bule, P, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Cell-surface Attachment of Bacterial Multienzyme Complexes Involves Highly Dynamic Protein-Protein Anchors.
J. Biol. Chem., 290, 2015
|
|
8AJY
 
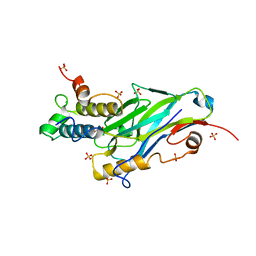 | | Ruminococcus flavefaciens Cohesin-Dockerin structure: dockerin from ScaH adaptor scaffoldin in complex with the cohesin from ScaE anchoring scaffoldin | | Descriptor: | CALCIUM ION, Cell-wall anchoring protein, Dockerin from ScaH, ... | | Authors: | Alves, V.D, Bule, P, Fontes, C.M.G.A, Carvalho, A.L.M, Najmudin, S, Duarte, M. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-function studies can improve binding affinity of cohesin-dockerin interactions for multi-protein assemblies.
Int.J.Biol.Macromol., 224, 2023
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOS
 
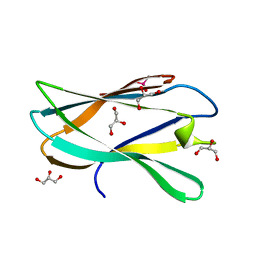 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5A6L
 
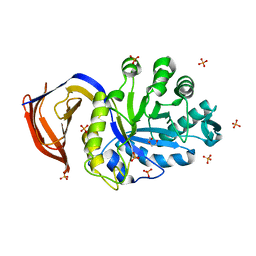 | | High resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with two xylobiose units bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5A6M
 
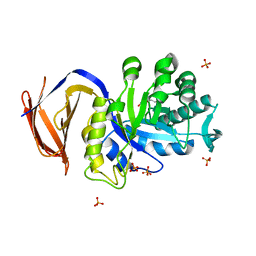 | | Determining the specificities of the catalytic site from the very high resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with a xylotetraose bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7R1N
 
 | | Crystal structure of the Tetrameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Paenibacillus illinoisensis IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
7R3T
 
 | | Crystal structure of the Dimeric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-1,3-glucanase bglH, CHLORIDE ION, ... | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Paenibacillus illinoisensis IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
5LXV
 
 | |
1MM0
 
 | | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger | | Descriptor: | Termicin | | Authors: | Da Silva, P, Jouvensal, L, Lamberty, M, Bulet, P, Caille, A, Vovelle, F. | | Deposit date: | 2002-09-02 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger
PROTEIN SCI., 12, 2003
|
|
1I2U
 
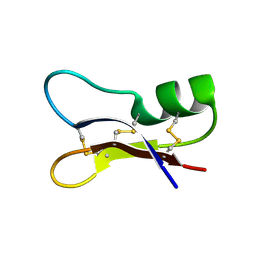 | | NMR SOLUTION STRUCTURES OF ANTIFUNGAL HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
1KFP
 
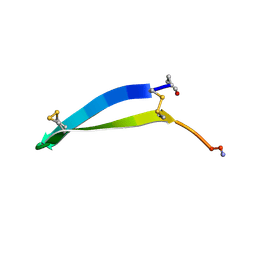 | | Solution structure of the antimicrobial 18-residue gomesin | | Descriptor: | GOMESIN | | Authors: | Mandard, N, Bulet, P, Caille, A, Daffre, S, Vovelle, F. | | Deposit date: | 2001-11-22 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider.
Eur.J.Biochem., 269, 2002
|
|
1UT3
 
 | | Solution Structure of Spheniscin-2, a beta-Defensin from Penguin Stomach Preserving Food | | Descriptor: | SPHENISCIN-2 | | Authors: | Landon, C, Labbe, H, Thouzeau, C, Bulet, P, Vovelle, F. | | Deposit date: | 2003-12-02 | | Release date: | 2004-05-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Spheniscin, a {Beta}-Defensin from the Penguin Stomach
J.Biol.Chem., 279, 2004
|
|
