4H6P
 
 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a R101A substitution. | | 分子名称: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | 著者 | Zhang, Y, Robinson, H, Buchko, G.W. | | 登録日 | 2012-09-19 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.556 Å) | | 主引用文献 | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | 分子名称: | De novo mini protein HHH_06 | | 著者 | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | 登録日 | 2016-04-22 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2O5F
 
 | | Crystal Structure of DR0079 from Deinococcus radiodurans at 1.9 Angstrom Resolution | | 分子名称: | Putative Nudix hydrolase DR_0079 | | 著者 | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H. | | 登録日 | 2006-12-05 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Functional and Structural Characterization of DR_0079 from Deinococcus radiodurans, a Novel Nudix Hydrolase with a Preference for Cytosine (Deoxy)ribonucleoside 5'-Di- and Triphosphates.
Biochemistry, 47, 2008
|
|
2O6W
 
 | | Crystal Structure of a Pentapeptide Repeat Protein (Rfr23) from the cyanobacterium Cyanothece 51142 | | 分子名称: | ARSENIC, Repeat Five Residue (Rfr) protein or pentapeptide repeat protein | | 著者 | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H, Pakrasi, H.B. | | 登録日 | 2006-12-08 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insights into the structural variation between pentapeptide repeat proteins-Crystal structure of Rfr23 from Cyanothece 51142.
J.Struct.Biol., 162, 2008
|
|
2ND3
 
 | | Solution structure of the de novo mini protein gEEH_04 | | 分子名称: | De novo mini protein EEH_04 | | 著者 | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | 登録日 | 2016-04-22 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
3S2Y
 
 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | 分子名称: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | 登録日 | 2011-05-17 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.244 Å) | | 主引用文献 | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
3GMT
 
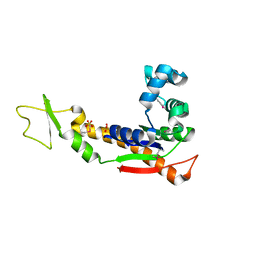 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | 分子名称: | Adenylate kinase, SULFATE ION | | 著者 | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2009-03-15 | | 公開日 | 2009-06-02 | | 最終更新日 | 2013-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
4E98
 
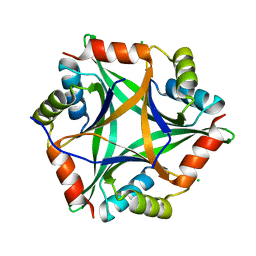 | |
4F83
 
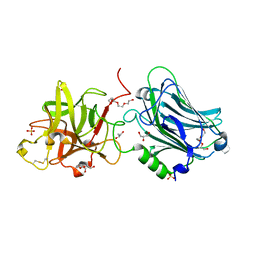 | | Crystal structure of the receptor binding domain of botulinum neurotoxin mosaic serotype C/D with a tetraethylene glycol molecule bound on the Hcn sub-domain and a sulfate ion at the putative active site | | 分子名称: | GLYCEROL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Zhang, Y, Buchko, G.W, Gardberg, A, Edwards, T.E, Sankaran, B, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-05-16 | | 公開日 | 2012-06-20 | | 最終更新日 | 2013-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into the functional role of the Hcn sub-domain of the receptor-binding domain of the botulinum neurotoxin mosaic serotype C/D.
Biochimie, 95, 2013
|
|
3OGG
 
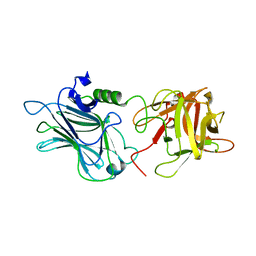 | | Crystal structure of the receptor binding domain of botulinum neurotoxin D | | 分子名称: | Botulinum neurotoxin type D | | 著者 | Zhang, Y, Gao, X, Qin, L, Buchko, G.W, Robinson, H, Varnum, S.M. | | 登録日 | 2010-08-16 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Structural analysis of the receptor binding domain of botulinum neurotoxin serotype D.
Biochem.Biophys.Res.Commun., 401, 2010
|
|
3PME
 
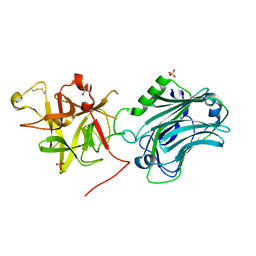 | | Crystal structure of the receptor binding domain of botulinum neurotoxin C/D mosaic serotype | | 分子名称: | GLYCEROL, SULFATE ION, Type C neurotoxin | | 著者 | Zhang, Y, Buchko, G.W, Qin, L, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2010-11-16 | | 公開日 | 2010-12-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Crystal structure of the receptor binding domain of the botulinum C-D mosaic neurotoxin reveals potential roles of lysines 1118 and 1136 in membrane interactions.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
3NJ2
 
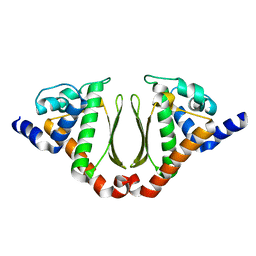 | | Crystal structure of cce_0566 from the cyanobacterium Cyanothece 51142, a protein associated with nitrogen fixation from the DUF269 family | | 分子名称: | DUF269-containing protein | | 著者 | Robinson, H, Ralston, C.Y, Addlagatta, A, Buchko, G.W. | | 登録日 | 2010-06-16 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal structure of cce_0566 from Cyanothece 51142, a protein associated with nitrogen fixation in the DUF269 family.
Febs Lett., 586, 2012
|
|
