4GYH
 
 | |
4H1I
 
 | |
3TZR
 
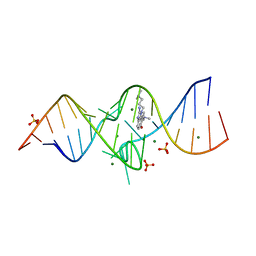 | | Structure of a Riboswitch-like RNA-ligand complex from the Hepatitis C Virus Internal Ribosome Entry Site | | Descriptor: | (8R)-8-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-1,7,8,9-tetrahydrochromeno[5,6-d]imidazol-2-amine, 5'-R(*CP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*CP*C)-3', 5'-R(*GP*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*UP*CP*GP*G)-3', ... | | Authors: | Dibrov, S.M, Ding, K, Brunn, N, Parker, M.A, Bergdahl, B.M, Wyles, D.L, Hermann, T. | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structure of a Riboswitch in the Hepatitis C Virus Internal Ribosome Entry Site
To be Published
|
|
7ZU0
 
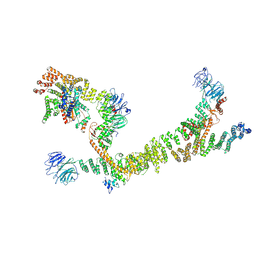 | | HOPS tethering complex from yeast | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar morphogenesis protein 6, ... | | Authors: | Shvarev, D, Schoppe, J, Koenig, C, Perz, A, Fuellbrunn, N, Kiontke, S, Langemeyer, L, Januliene, D, Schnelle, K, Kuemmel, D, Froehlich, F, Moeller, A, Ungermann, C. | | Deposit date: | 2022-05-11 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery.
Elife, 11, 2022
|
|
1KY8
 
 | | Crystal Structure of the Non-phosphorylating glyceraldehyde-3-phosphate Dehydrogenase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pohl, E, Brunner, N, Wilmanns, M, Hensel, R. | | Deposit date: | 2002-02-04 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Allosteric Non-phosphorylating
glyceraldehyde-3-phosphate Dehydrogenase from the Hyperthermophilic
Archaeum Thermoproteus tenax
J.Biol.Chem., 277, 2002
|
|
