6VJG
 
 | | Csx3-I222 Crystal Form at 1.8 Angstrom Resolution | | Descriptor: | CRISPR-associated protein, Csx3 family | | Authors: | Brown, S, Charbonneau, A, Burman, N, Gauvin, C.C, Lawrence, C.M. | | Deposit date: | 2020-01-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA 4 .
J.Biol.Chem., 295, 2020
|
|
176D
 
 | |
4JVA
 
 | | Crystal Structure of RIIbeta(108-402) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
3IDB
 
 | | Crystal structure of (108-268)RIIb:C holoenzyme of cAMP-dependent protein kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Brown, S.H.J, Wu, J, Kim, C, Alberto, K, Taylor, S.S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures.
J.Mol.Biol., 393, 2009
|
|
3IDC
 
 | | Crystal structure of (102-265)RIIb:C holoenzyme of cAMP-dependent protein kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Brown, S.H.J, Wu, J, Kim, C, Alberto, K, Taylor, S.S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures.
J.Mol.Biol., 393, 2009
|
|
4JV4
 
 | | Crystal Structure of RIalpha(91-379) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
1BEB
 
 | |
4TGF
 
 | | SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA | | Descriptor: | DES-VAL-1,VAL-2,TRANSFORMING GROWTH FACTOR, ALPHA | | Authors: | Kline, T.P, Brown, F.K, Brown, S.C, Jeffs, P.W, Kopple, K.D, Mueller, L. | | Deposit date: | 1990-06-13 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human transforming growth factor alpha derived from 1H NMR data.
Biochemistry, 29, 1990
|
|
8E1F
 
 | | Sterile Alpha Motif Domain of Human Translocation ETS Leukemia, Non-Polymer Crystal Form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wilson, E, Nawarathnage, S, Stewart, C, Brown, S, Bezzant, B, Bunn, D, Towne, N, Pedroza Romo, M.J, Moody, J.D. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Sterile Alpha Motif Domain of Human Translocation ETS Leukemia, Non-Polymer Crystal Form
To Be Published
|
|
8U1H
 
 | | Axle-less Bacillus sp. PS3 F1 ATPase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Furlong, E.J, Zeng, Y.C, Brown, S.H.J, Sobti, M, Stewart, A.G. | | Deposit date: | 2023-09-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The molecular structure of an axle-less F 1 -ATPase.
Biochim Biophys Acta Bioenerg, 2024
|
|
8U0N
 
 | | Crystal structure of isopentenyl phosphate kinase from Thermococcus paralvinellae bound to 2-cyclopentylideneethyl monophosphate and ADP | | Descriptor: | 2-cyclopentylideneethyl dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, Isopentenyl phosphate kinase | | Authors: | Singh, S, Thomas, L.M, Johnson, B.P, Brown, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ternary complexes of isopentenyl phosphate kinase from Thermococcus paralvinellae reveal molecular determinants of non-natural substrate specificity.
Proteins, 92, 2024
|
|
8U0L
 
 | | Crystal structure of isopentenyl phosphate kinase from Thermococcus paralvinellae bound to (E)-But-2-en-1-yl monophosphate and ADP | | Descriptor: | (2E)-but-2-en-1-yl dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, Isopentenyl phosphate kinase | | Authors: | Singh, S, Thomas, L.M, Johnson, B.P, Brown, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ternary complexes of isopentenyl phosphate kinase from Thermococcus paralvinellae reveal molecular determinants of non-natural substrate specificity.
Proteins, 92, 2024
|
|
8U0K
 
 | | Crystal structure of isopentenyl phosphate kinase from Thermococcus paralvinellae bound to DMAP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dimethylallyl monophosphate, Isopentenyl phosphate kinase | | Authors: | Singh, S, Thomas, L.M, Johnson, B.P, Brown, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ternary complexes of isopentenyl phosphate kinase from Thermococcus paralvinellae reveal molecular determinants of non-natural substrate specificity.
Proteins, 92, 2024
|
|
8U0M
 
 | | Crystal structure of isopentenyl phosphate kinase from Thermococcus paralvinellae bound to (E)-2-methylbut-2-en-1-yl monophosphate and ATP | | Descriptor: | (2E)-2-methylbut-2-en-1-yl dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Singh, S, Thomas, L.M, Johnson, B.P, Brown, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Ternary complexes of isopentenyl phosphate kinase from Thermococcus paralvinellae reveal molecular determinants of non-natural substrate specificity.
Proteins, 92, 2024
|
|
1OXW
 
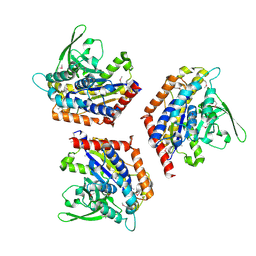 | | The Crystal Structure of SeMet Patatin | | Descriptor: | Patatin | | Authors: | Rydel, T.J, Williams, J.M, Krieger, E, Moshiri, F, Stallings, W.C, Brown, S.M, Pershing, J.C, Purcell, J.P, Alibhai, M.F. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure, Mutagenesis, and Activity Studies Reveal that Patatin Is a
Lipid Acyl Hydrolase with a Ser-Asp Catalytic Dyad
Biochemistry, 42, 2003
|
|
7T8J
 
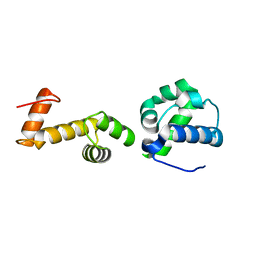 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a GSGG linker | | Descriptor: | CHLORIDE ION, Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Soleimani, S, Pedroza Romo, M.J, Smith, T, Brown, S, Doukov, T, Moody, J.D. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
1JWE
 
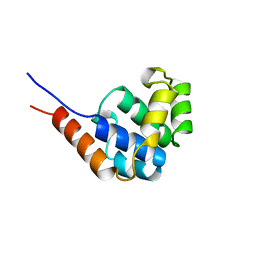 | | NMR Structure of the N-Terminal Domain of E. Coli Dnab Helicase | | Descriptor: | PROTEIN (DNAB HELICASE) | | Authors: | Weigelt, J, Brown, S.E, Miles, C.S, Dixon, N.E, Otting, G. | | Deposit date: | 1999-01-22 | | Release date: | 1999-01-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer.
Structure Fold.Des., 7, 1999
|
|
1J54
 
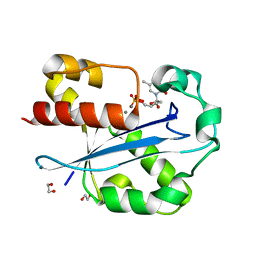 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1J53
 
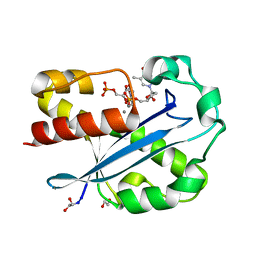 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1TBO
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, 30 STRUCTURES | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
1VYX
 
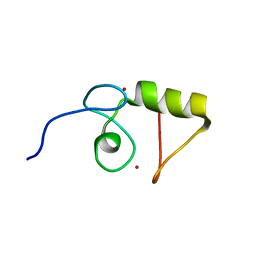 | | Solution structure of the KSHV K3 N-terminal domain | | Descriptor: | ORF K3, ZINC ION | | Authors: | Dodd, R.B, Allen, M.D, Brown, S.E, Sanderson, C.M, Duncan, L.M, lehner, P.J, Bycroft, M, Read, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-10-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Kaposi'S Sarcoma-Associated Herpesvirus K3 N-Terminal Domain Reveals a Novel E2-Binding C4Hc3-Type Ring Domain
J.Biol.Chem., 279, 2004
|
|
1TBN
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
1RL3
 
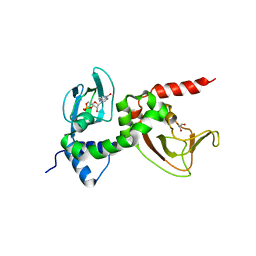 | | Crystal structure of cAMP-free R1a subunit of PKA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Brown, S, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2003-11-24 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B.
Structure, 12, 2004
|
|
2QVS
 
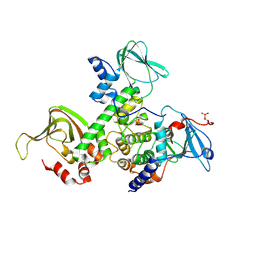 | | Crystal Structure of Type IIa Holoenzyme of cAMP-dependent Protein Kinase | | Descriptor: | cAMP-dependent protein kinase type II-alpha regulatory subunit, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Wu, J, Brown, S.H.J, von Daake, S, Taylor, S.S. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PKA type IIalpha holoenzyme reveals a combinatorial strategy for isoform diversity.
Science, 318, 2007
|
|
8FZ4
 
 | | The von Willebrand factor A domain of Anthrax toxin receptor 2 | | Descriptor: | Anthrax toxin receptor 2, CHLORIDE ION, MAGNESIUM ION | | Authors: | Pedroza-Romo, M.J, Soleimani, S, Lewis, A, Smith, T, Brown, S, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
