4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
3DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
2DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1T90
 
 | | Crystal structure of methylmalonate semialdehyde dehydrogenase from Bacillus subtilis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable methylmalonate-semialdehyde dehydrogenase | | Authors: | Dubourg, H, Didierjean, C, Stines-Chaumeil, C, Talfournier, F, Branlant, G, Aubry, A, Corbier, C. | | Deposit date: | 2004-05-14 | | Release date: | 2006-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of Methylmalonate-Semialdehyde Dehydrogenase from Bacillus subtilis.
To be published
|
|
1GAE
 
 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1GAD
 
 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
3MMH
 
 | | X-ray structure of free methionine-R-sulfoxide reductase from neisseria meningitidis in complex with its substrate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Gruez, A, Libiad, M, Boschi-Muller, S, Branlant, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Free Methionine-R-sulfoxide Reductase from Neisseria meningitidis.
J.Biol.Chem., 285, 2010
|
|
4LRT
 
 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, COENZYME A, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
4LRS
 
 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
1CF2
 
 | | THREE-DIMENSIONAL STRUCTURE OF D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM THE HYPERTHERMOPHILIC ARCHAEON METHANOTHERMUS FERVIDUS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Charron, C, Talfournier, F, Isuppov, M.N, Branlant, G, Littlechild, J.A, Vitoux, B, Aubry, A. | | Deposit date: | 1999-03-24 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of D-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanothermus fervidus.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2QE0
 
 | | Thioacylenzyme Intermediate of GAPN from S. Mutans, New Data Integration and Refinement. | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Corbier, C, Didierjean, C, Bricogne, G, Branlant, G, D'Ambrosio, K, Vonrhein, C. | | Deposit date: | 2007-06-22 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The First Crystal Structure of a Thioacylenzyme Intermediate in the ALDH Family: New Coenzyme Conformation and Relevance to Catalysis
Biochemistry, 45, 2006
|
|
2XF8
 
 | | Structure of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli in complex with a NAD cofactor analog (3-Chloroacetyl adenine pyridine dinucleotide) and sulfate anion | | Descriptor: | 3-(CHLOROACETYL) PYRIDINE ADENINE DINUCLEOTIDE, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2X5K
 
 | | Structure of an active site mutant of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, ... | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2X5J
 
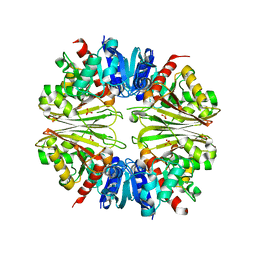 | | Crystal structure of the Apoform of the D-Erythrose-4-phosphate dehydrogenase from E. coli | | Descriptor: | D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, PHOSPHATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
1EUH
 
 | | APO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE FROM STREPTOCOCCUS MUTANS | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
1FF3
 
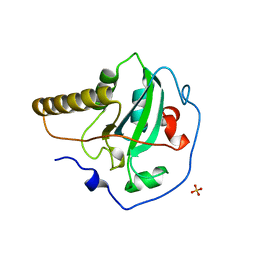 | | STRUCTURE OF THE PEPTIDE METHIONINE SULFOXIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | PEPTIDE METHIONINE SULFOXIDE REDUCTASE, SULFATE ION | | Authors: | Tete-Favier, F, Cobessi, D, Boschi-Muller, S, Azza, S, Branlant, G, Aubry, A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Escherichia coli peptide methionine sulphoxide reductase at 1.9 A resolution.
Structure Fold.Des., 8, 2000
|
|
1NQO
 
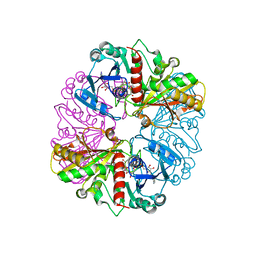 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate
Dehydrogenase from Bacillus stearothermophilus with NAD and
D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQA
 
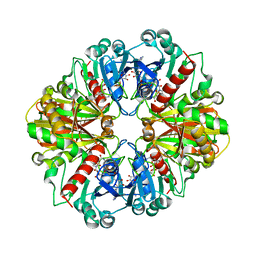 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ala Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating
Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus
with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQ5
 
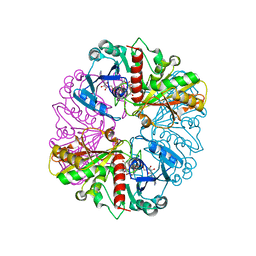 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NPT
 
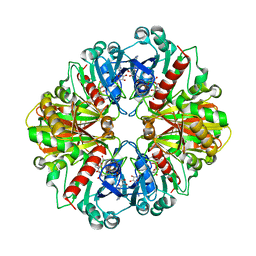 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 replaced by Ala complexed with NAD+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1QI6
 
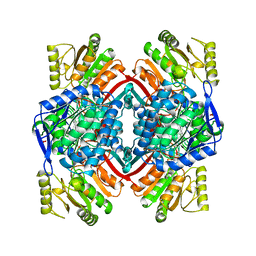 | | SECOND APO FORM OF AN NADP DEPENDENT ALDEHYDE DEHYDROGENASE WITH GLU250 SITUATED 3.7 A FROM CYS284 | | Descriptor: | PROTEIN (NADP DEPENDENT NONPHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1999-06-02 | | Release date: | 2001-01-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 300, 2000
|
|
1QI1
 
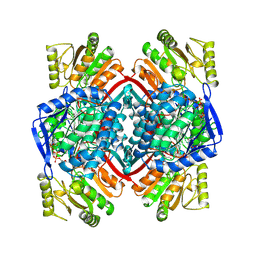 | | Ternary Complex of an NADP Dependent Aldehyde Dehydrogenase | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (NADP-DEPENDENT NONPHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), ... | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1999-06-02 | | Release date: | 2001-01-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 300, 2000
|
|
2ID2
 
 | | GAPN T244S mutant X-ray structure at 2.5 A | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pailot, A, D'Ambrosio, K, Corbier, C, Talfournier, F, Branlant, G. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Invariant Thr(244) is essential for the efficient acylation step of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans.
Biochem.J., 400, 2006
|
|
2EUH
 
 | | HOLO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE COMPLEX WITH NADP+ | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
