8P6G
 
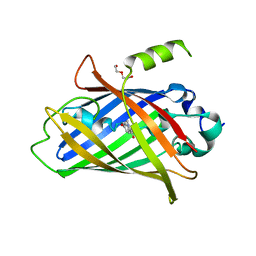 | | Crystal structure of the improved version of the Genetically Encoded Green Calcium Indicator YTnC2-5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Genetically Encoded Green Calcium Indicator YTnC2-5 based on Troponin C from toadfish | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Popov, V.O, Subach, F.V. | | Deposit date: | 2023-05-26 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | YTnC2, an improved genetically encoded green calcium indicator based on toadfish troponin C.
Febs Open Bio, 13, 2023
|
|
8P1H
 
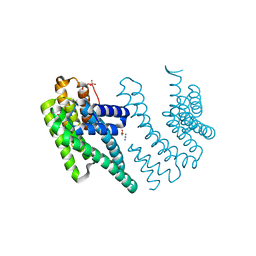 | | Crystal structure of the chimera of human 14-3-3 zeta and phosphorylated cytoplasmic loop fragment of the alpha7 acetylcholine receptor | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, BENZOIC ACID, ... | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Lyukmanova, E.N, Sluchanko, N.N. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure reveals canonical recognition of the phosphorylated cytoplasmic loop of human alpha7 nicotinic acetylcholine receptor by 14-3-3 protein.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
8PYH
 
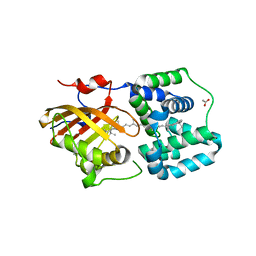 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Crinalium epipsammum PCC 9333 | | Descriptor: | ACETATE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | Deposit date: | 2023-07-25 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8PZK
 
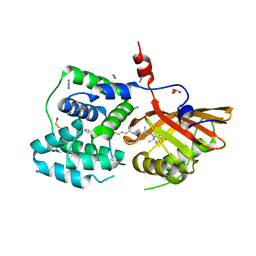 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
2ZO5
 
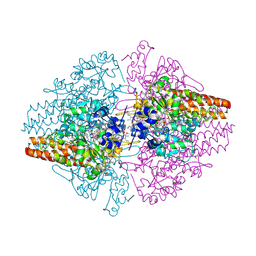 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with azide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
6ZL7
 
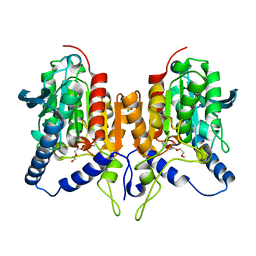 | | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, MAGNESIUM ION, PMGL2 | | Authors: | Goryaynova, D.A, Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY
To Be Published
|
|
4WWV
 
 | | Aminopeptidase APDkam598 from the archaeon Desulfurococcus kamchatkensis | | Descriptor: | Aminopeptidase from family M42 | | Authors: | Petrova, T, Boyko, K.M, Rakitina, T.V, Korzhenevskiy, D.A, Gorbacheva, M.A, Popov, V.O. | | Deposit date: | 2014-11-12 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structure of the dodecamer of the aminopeptidase APDkam598 from the archaeon Desulfurococcus kamchatkensis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8PNX
 
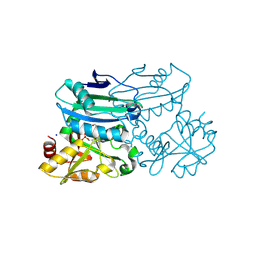 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in PMP form | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of dual substrate recognition in the transaminase from Blastococcus saxobsidens specific to D-amino acids and R-amines
To Be Published
|
|
2OT4
 
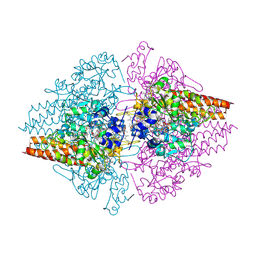 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
5EK6
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 complexed with NADP and isobutyraldehyde | | Descriptor: | 2-methylpropanal, Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Polyakov, K.M, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EXF
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5F2C
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-12-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EUY
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EEB
 
 | | Apo form of thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 | | Descriptor: | Aldehyde dehydrogenase | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-10-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.038 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EKC
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Shabalin, I.G, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+
To Be Published
|
|
8OSI
 
 | | Genetically encoded green ratiometric calcium indicator FNCaMP in calcium-bound state | | Descriptor: | CALCIUM ION, mNeonGreen,Calmodulin,Protein kinase domain-containing protein, {(4Z)-2-(aminomethyl)-4-[(4-hydroxyphenyl)methylidene]-5-oxo-4,5-dihydro-1H-imidazol-1-yl}acetic acid | | Authors: | Varfolomeeva, L.A, Boyko, K.M, Nikolaeva, A.Y, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | FNCaMP, ratiometric green calcium indicator based on mNeonGreen protein.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
8OQ0
 
 | | Crystal structure of tailspike depolymerase (APK09_gp48) from Acinetobacter phage APK09 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tailspike protein | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Popova, A.V, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
8OPZ
 
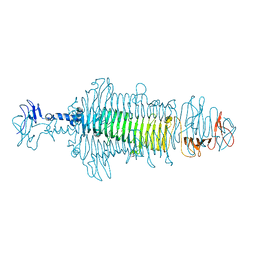 | | Crystal structure of a tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Descriptor: | GLYCEROL, Tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
8OQ1
 
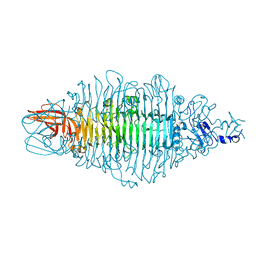 | | Crystal structure of tailspike depolymerase (APK14_gp49) from Acinetobacter phage vB_AbaP_APK14 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii.
Int J Mol Sci, 24, 2023
|
|
8BPN
 
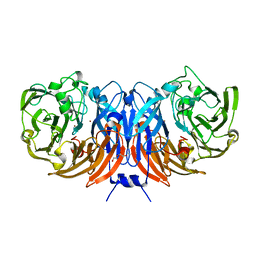 | | The structure of thiocyanate dehydrogenase mutant form with Phe 436 replaced by Gln from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Twin-arginine translocation signal domain-containing protein | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Kulikova, O.G, Dergousova, N.I, Rakitina, T.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-01-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Probing the Role of a Conserved Phenylalanine in the Active Site of Thiocyanate Dehydrogenase
Crystals, 12, 2022
|
|
3RR5
 
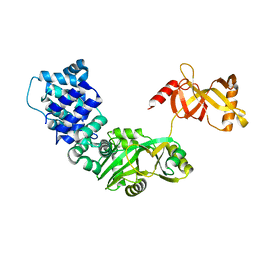 | | DNA ligase from the archaeon Thermococcus sp. 1519 | | Descriptor: | DNA ligase, MAGNESIUM ION | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Popov, V.O, Polyakov, K.M, Ravin, N.V, Shabalin, I.G, Skryabin, K.G, Stekhanova, T.N, Kovalchuk, M.V. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.018 Å) | | Cite: | ATP-dependent DNA ligase from Thermococcus sp. 1519 displays a new arrangement of the OB-fold domain.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3SCE
 
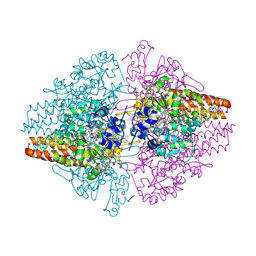 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with a covalent bond between the CE1 atom of Tyr303 and the CG atom of Gln360 (TvNiRb) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8ONO
 
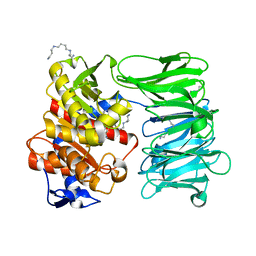 | | Modified oligopeptidase B from S. proteamaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution
To Be Published
|
|
5JFQ
 
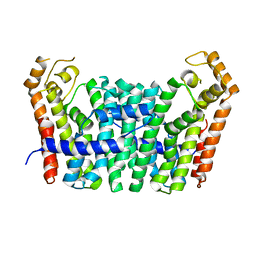 | | Geranylgeranyl Pyrophosphate Synthetase from archaeon Geoglobus acetivorans | | Descriptor: | Geranylgeranyl Pyrophosphate Synthetase | | Authors: | Petrova, T, Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of geranylgeranyl pyrophosphate synthase GACE1337 from the hyperthermophilic archaeon Geoglobus acetivorans.
Extremophiles, 22, 2018
|
|
4RGZ
 
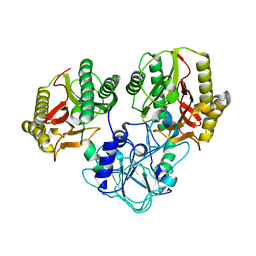 | | Crystal structure of recombinant prolidase from Thermococcus sibiricus at P21221 spacegroup | | Descriptor: | PHOSPHATE ION, Xaa-Pro aminopeptidase, ZINC ION | | Authors: | Timofeev, V.I, Korgenevsky, D.A, Gorbacheva, M.A, Boyko, K.M, Slutsky, E, Rakitina, T.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2014-10-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
