8EIW
 
 | | Cobalt(II)-substituted Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
8EIY
 
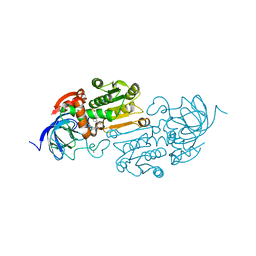 | | Cobalt(II)-substituted S48T Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
8EIX
 
 | | Cobalt(II)-substituted S48A Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
7SJJ
 
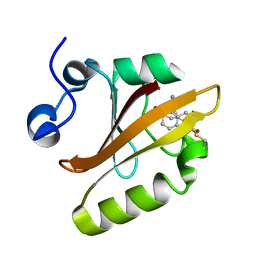 | | Crystal structure of photoactive yellow protein (PYP); F96oCNF construct | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | Deposit date: | 2021-10-17 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
8EE3
 
 | |
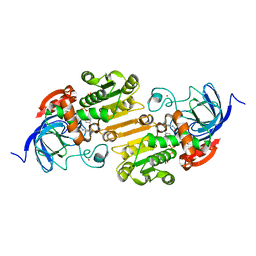
8ECT
 
 | |
8ECU
 
 | |
8ECS
 
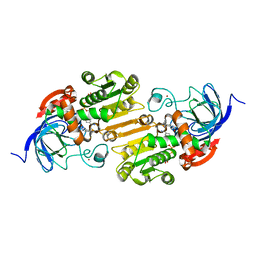 | |
7SPV
 
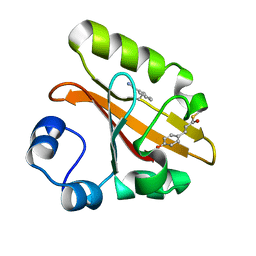 | | Crystal structure of photoactive yellow protein (PYP); F92oCNF construct | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
7SPW
 
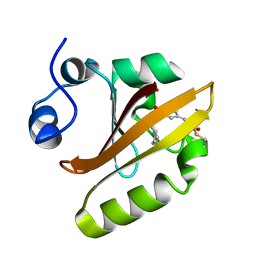 | | Crystal structure of photoactive yellow protein (PYP); F62oCNF construct | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
7SPX
 
 | | Crystal structure of photoactive yellow protein (PYP); F28oCNF construct | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
7U6Q
 
 | | TEM-1 beta-lactamase | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-03-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
3OR0
 
 | |
7MH9
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-nitrotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH5
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-iodotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH4
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-bromotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH8
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-methyltyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH3
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-chlorotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J.B, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3OQY
 
 | |
3OQZ
 
 | |
7RM6
 
 | |
6PFU
 
 | |
6PFR
 
 | |
6PFS
 
 | |
6PFT
 
 | |
