2V5D
 
 | | Structure of a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. | | Descriptor: | CALCIUM ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2V8I
 
 | |
2W47
 
 | | Clostridium thermocellum CBM35 in complex with delta-4,5- anhydrogalacturonic acid | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid, CALCIUM ION, LIPOLYTIC ENZYME, ... | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VZT
 
 | | Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine | | Descriptor: | 4-nitrophenyl 2-amino-2-deoxy-beta-D-glucopyranoside, ACETATE ION, CADMIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2W87
 
 | | Xyl-CBM35 in complex with glucuronic acid containing disaccharide. | | Descriptor: | CALCIUM ION, ESTERASE D, UNKNOWN LIGAND, ... | | Authors: | Montainer, C, Bueren, A.L.v, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VZQ
 
 | |
2VZV
 
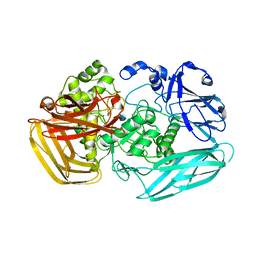 | | Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, EXO-BETA-D-GLUCOSAMINIDASE | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2W46
 
 | | CBM35 from Cellvibrio japonicus Abf62 | | Descriptor: | CALCIUM ION, ESTERASE D, SODIUM ION | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W1U
 
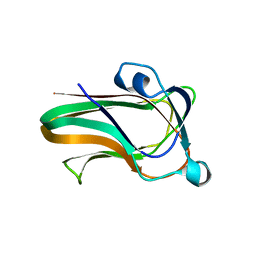 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ACETATE ION, ... | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
2VZP
 
 | |
2VZS
 
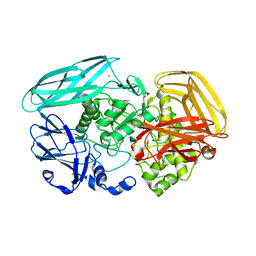 | | Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, CADMIUM ION, EXO-BETA-D-GLUCOSAMINIDASE, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2VZR
 
 | |
2VZU
 
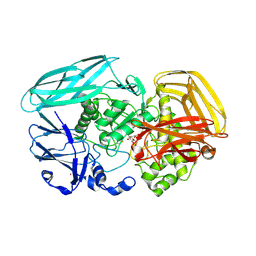 | | Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine | | Descriptor: | 4-nitrophenyl 2-amino-2-deoxy-beta-D-glucopyranoside, ACETATE ION, CADMIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2W1Q
 
 | |
2W1S
 
 | |
2VNR
 
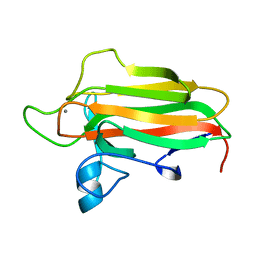 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens. | | Descriptor: | CALCIUM ION, CPE0329 | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
2W91
 
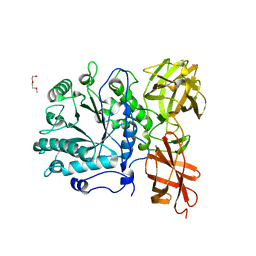 | | Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D. | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE D, PENTAETHYLENE GLYCOL | | Authors: | Abbott, D.W, Macauley, M.S, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Streptococcus Pneumoniae Endohexosaminidase D, Structural and Mechanistic Insight Into Substrate-Assisted Catalysis in Family 85 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2W92
 
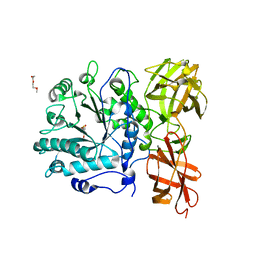 | | Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D, in complex with NAG-thiazoline. | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE D, PENTAETHYLENE GLYCOL | | Authors: | Abbott, D.W, Macauley, M.S, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Streptococcus Pneumoniae Endohexosaminidase D, Structural and Mechanistic Insight Into Substrate-Assisted Catalysis in Family 85 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2VO8
 
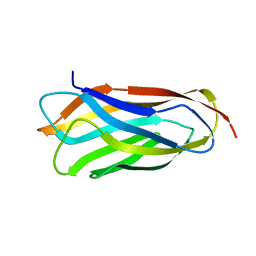 | | Cohesin module from Clostridium perfringens ATCC13124 family 33 glycoside hydrolase. | | Descriptor: | EXO-ALPHA-SIALIDASE | | Authors: | Gregg, K, Adams, J.J, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Clostridium Perfringens Toxin Complex Formation.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6XJ9
 
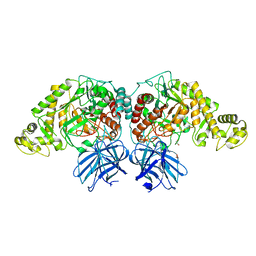 | | Structure of PfGH50B | | Descriptor: | 1,2-ETHANEDIOL, Agarase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-06-23 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of PfGH50B, an agarase from the marine bacterium Pseudoalteromonas fuliginea PS47
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6XSZ
 
 | |
6B0K
 
 | | Crystal structure of Ps i-CgsB C78S in complex with k-carrapentaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-D-galactose, 4-O-sulfo-beta-D-galactopyranose, ... | | Authors: | Hettle, A, Boraston, A.B. | | Deposit date: | 2017-09-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6DRR
 
 | | Crystal structure of Cj0485 dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Short-chain dehydrogenase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2018-06-12 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus.
Commun Biol, 3, 2020
|
|
6DS1
 
 | |
6DNS
 
 | | Endo-fucoidan hydrolase MfFcnA9 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
