3ZM0
 
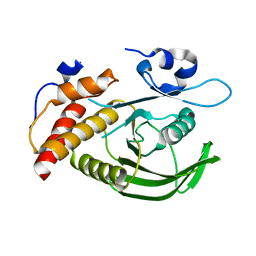 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM2
 
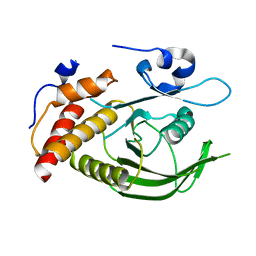 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM3
 
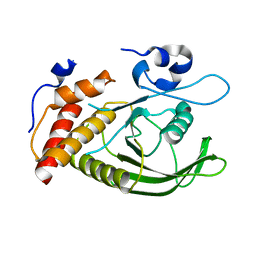 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM1
 
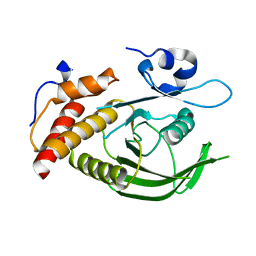 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
4A11
 
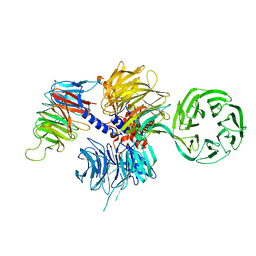 | | Structure of the hsDDB1-hsCSA complex | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | Authors: | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
2WNI
 
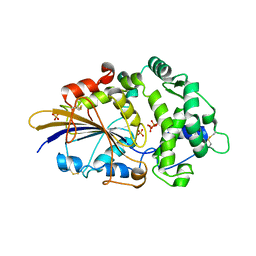 | |
2WU0
 
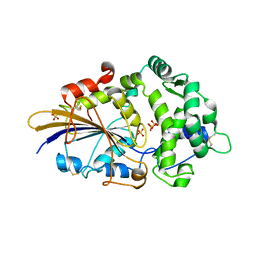 | |
2WNH
 
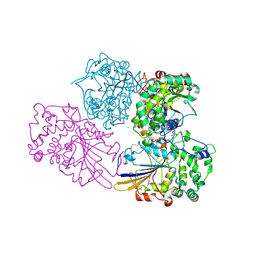 | | Crystal Structure Analysis of Klebsiella sp ASR1 Phytase | | Descriptor: | 3-PHYTASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bohm, K, Mueller, J.J, Heinemann, U. | | Deposit date: | 2009-07-09 | | Release date: | 2010-04-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Klebsiella Sp. Asr1 Phytase Suggests Substrate Binding to a Preformed Active Site that Meets the Requirements of a Plant Rhizosphere Enzyme.
FEBS J., 277, 2010
|
|
