5UXZ
 
 | |
6XCV
 
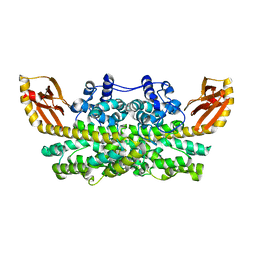 | |
8CVD
 
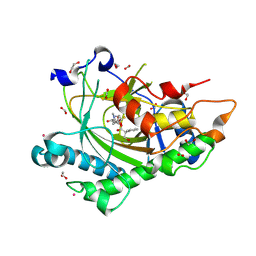 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and scopolamine | | Descriptor: | (1R,2R,4S,5S,7s)-9-methyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-7-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structure of the H6H cyclization product complex
To Be Published
|
|
8CVA
 
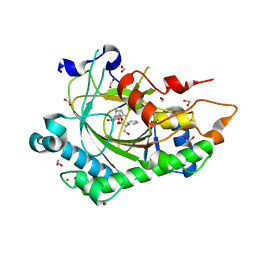 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure of the H6H hydroxylation product complex
To Be Published
|
|
8CVC
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure mimicking the H6H cyclization ferryl complex
To Be Published
|
|
8CV8
 
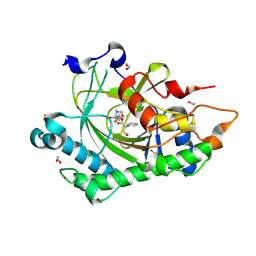 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the H6H hydroxylation reactant complex
To Be Published
|
|
8CVB
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the H6H cyclization reactant complex
To Be Published
|
|
8CV9
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the H6H hydroxylation reactant complex
To Be Published
|
|
8CVG
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of the L289F H6H cyclization reactant complex
To Be Published
|
|
8CVF
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the L289F H6H ferryl-mimicking complex
To Be Published
|
|
8CVE
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of the L289F H6H hydroxylation reactant complex
To Be Published
|
|
8CVH
 
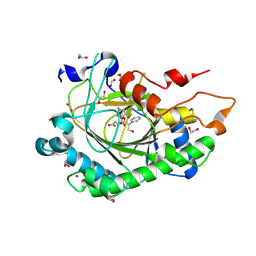 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the L289F H6H cyclization ferryl-mimicking complex
To Be Published
|
|
5HR6
 
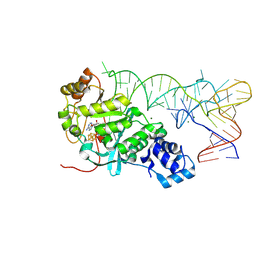 | | X-ray crystal structure of C118A RlmN with cross-linked tRNA purified from Escherichia coli | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Schwalm, E.L, Grove, T.L, Booker, S.J, Boal, A.K. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-13 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA.
Science, 352, 2016
|
|
5HR7
 
 | | X-ray crystal structure of C118A RlmN from Escherichia coli with cross-linked in vitro transcribed tRNA | | Descriptor: | 5'-DEOXYADENOSINE, Dual-specificity RNA methyltransferase RlmN, IRON/SULFUR CLUSTER, ... | | Authors: | Schwalm, E.L, Grove, T.L, Booker, S.J, Boal, A.K. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA.
Science, 352, 2016
|
|
5IQV
 
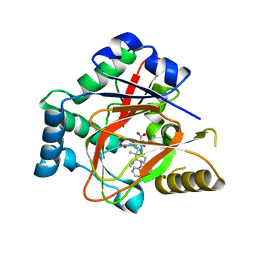 | | WelO5 bound to Fe, Cl, 2-oxoglutarate, 12-epifischerindole U, and nitric oxide | | Descriptor: | (6aS,9R,10R,10aS)-9-ethyl-10-isocyano-6,6,9-trimethyl-5,6,6a,7,8,9,10,10a-octahydroindeno[2,1-b]indole, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Mitchell, A.J, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
5IQT
 
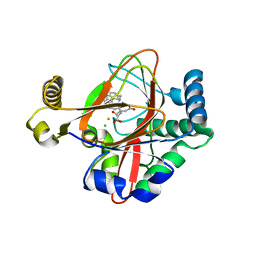 | | WelO5 bound to Fe(II), Cl, 2-oxoglutarate, and 12-epifischerindole U | | Descriptor: | (6aS,9R,10R,10aS)-9-ethyl-10-isocyano-6,6,9-trimethyl-5,6,6a,7,8,9,10,10a-octahydroindeno[2,1-b]indole, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Mitchell, A.J, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
5IQU
 
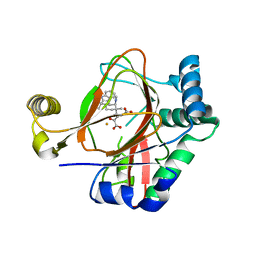 | | WelO5 G166D variant bound to Fe(II), 2-oxoglutarate, and 12-epifischerindole U | | Descriptor: | (6aS,9R,10R,10aS)-9-ethyl-10-isocyano-6,6,9-trimethyl-5,6,6a,7,8,9,10,10a-octahydroindeno[2,1-b]indole, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Mitchell, A.J, Maggiolo, A.O, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
5IQS
 
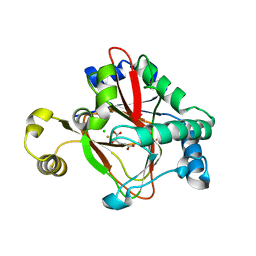 | | WelO5 bound to Fe(II), Cl, and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Mitchell, A.J, Ananth, N, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
6MP8
 
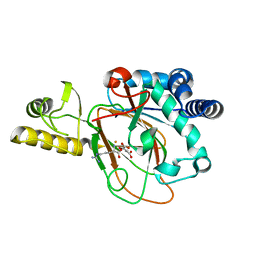 | | X-ray crystal structure of VioC bound to Fe(II), D-arginine, and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent L-arginine hydroxylase, D-ARGININE, ... | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2018-10-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | alpha-Amine Desaturation of d-Arginine by the Iron(II)- and 2-(Oxo)glutarate-Dependent l-Arginine 3-Hydroxylase, VioC.
Biochemistry, 57, 2018
|
|
6MP9
 
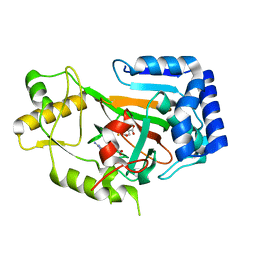 | | X-ray crystal structure of VioC bound to Fe(II), 2-oxo-5-guanidinopentanoic acid, and succinate | | Descriptor: | 5-carbamimidamido-2-oxopentanoic acid, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2018-10-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | alpha-Amine Desaturation of d-Arginine by the Iron(II)- and 2-(Oxo)glutarate-Dependent l-Arginine 3-Hydroxylase, VioC.
Biochemistry, 57, 2018
|
|
6NPC
 
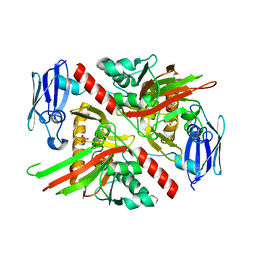 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe, 2OG, and 2-trimethylaminoethylphosphonate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, N,N,N-trimethyl-2-phosphonoethan-1-aminium, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6NPD
 
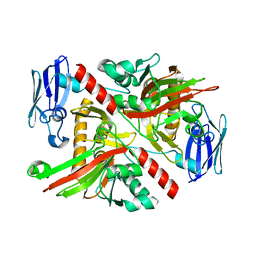 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe, 2OG, and (R)-1-hydroxy-2-trimethylaminoethylphosphonate | | Descriptor: | (2R)-2-hydroxy-N,N,N-trimethyl-2-phosphonoethan-1-aminium, FE (II) ION, TmpA, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6NPA
 
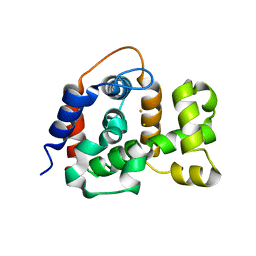 | | X-ray crystal structure of TmpB, (R)-1-hydroxy-2-trimethylaminoethylphosphonate oxygenase, with (R)-1-hydroxy-2-trimethylaminoethylphosphonate | | Descriptor: | (2R)-2-hydroxy-N,N,N-trimethyl-2-phosphonoethan-1-aminium, FE (II) ION, FE (III) ION, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6NPB
 
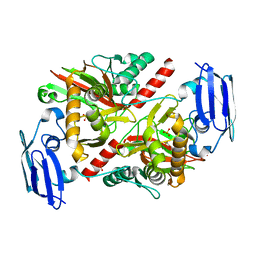 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, SULFATE ION, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6ONP
 
 | |
